7ONU
 
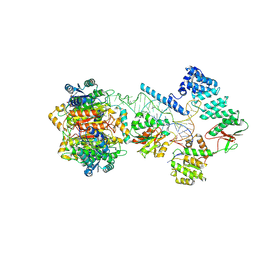 | | Structure of human mitochondrial RNase P in complex with mitochondrial pre-tRNA-Tyr | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, MAGNESIUM ION, Mitochondrial Precursor tRNA-Tyr, ... | | Authors: | Bhatta, A, Dienemann, C, Cramer, P, Hillen, H.S. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of RNA processing by human mitochondrial RNase P.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7E3X
 
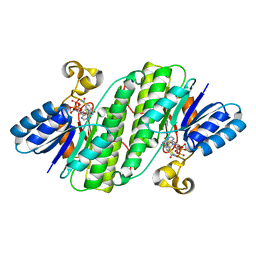 | | Crystal structure of SDR family NAD(P)-dependent oxidoreductase from exiguobacterium | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Chen, L, Tang, J, Yuan, S, Zhang, F, Chen, S. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-guided evolution of a ketoreductase forefficient and stereoselective bioreduction of bulkyalpha-aminobeta-keto esters
Catalysis Science And Technology, 2021
|
|
7E28
 
 | |
7E24
 
 | |
7BTM
 
 | | SDR protein/resistance protein NapW | | Descriptor: | Short chain dehydrogenase | | Authors: | Wen, W.H, Tang, G.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08311653 Å) | | Cite: | Reductive inactivation of the hemiaminal pharmacophore for resistance against tetrahydroisoquinoline antibiotics.
Nat Commun, 12, 2021
|
|
7BSX
 
 | | SDR protein NapW-NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Wen, W.H, Tang, G.L. | | Deposit date: | 2020-03-31 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reductive inactivation of the hemiaminal pharmacophore for resistance against tetrahydroisoquinoline antibiotics.
Nat Commun, 12, 2021
|
|
7E6O
 
 | |
7ULH
 
 | |
8DT1
 
 | |
8B5T
 
 | |
8B5U
 
 | |
7X3Z
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NAD | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, 17-beta-hydroxysteroid dehydrogenase type 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, T, Lin, S.X, Yin, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New insights into the substrate inhibition of human 17 beta-hydroxysteroid dehydrogenase type 1.
J.Steroid Biochem.Mol.Biol., 228, 2023
|
|
8CVN
 
 | | Cryo-EM Structure of Human 15-PGDH in Complex with Small Molecule SW209415 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 15-hydroxyprostaglandin dehydrogenase [NAD(+)], 2-[butyl(oxidanyl)-$l^{3}-sulfanyl]-4-(2,3-dimethylimidazol-4-yl)-6-(1,3-thiazol-2-yl)thieno[2,3-b]pyridin-3-amine | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Small molecule inhibitors of 15-PGDH exploit a physiologic induced-fit closing system.
Nat Commun, 14, 2023
|
|
8FD8
 
 | | human 15-PGDH with NADH bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2022-12-02 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Small molecule inhibitors of 15-PGDH exploit a physiologic induced-fit closing system.
Nat Commun, 14, 2023
|
|
7X5J
 
 | | ACP-dependent oxoacyl reductase | | Descriptor: | 3-oxoacyl-ACP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHENYLALANINE | | Authors: | Wang, S, Bai, L. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ACP-dependent oxoacyl reductase
To Be Published
|
|
8CWL
 
 | | Cryo-EM structure of Human 15-PGDH in complex with small molecule SW222746 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 15-hydroxyprostaglandin dehydrogenase [NAD(+)], 2-methyl-6-[7-(piperidine-1-carbonyl)quinoxalin-2-yl]isoquinolin-1(2H)-one | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-05-19 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Small molecule inhibitors of 15-PGDH exploit a physiologic induced-fit closing system.
Nat Commun, 14, 2023
|
|
8A7S
 
 | |
8AEW
 
 | |
8A30
 
 | |
8AEO
 
 | |
8AEQ
 
 | |
8AER
 
 | |
8A8T
 
 | |
8AET
 
 | |
8HI4
 
 | |
