4Q6I
 
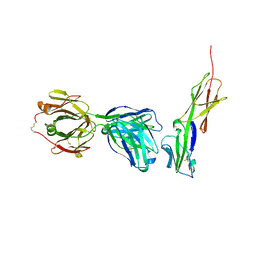 | | Crystal structure of murine 2D5 Fab, a potent anti-CD4 HIV-1-neutralizing antibody in complex with CD4 | | Descriptor: | Heavy chain of murine 2D5 Fab, Light chain of murine 2D5 Fab, T-cell surface glycoprotein CD4 | | Authors: | Boyington, J.C, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor.
Sci Transl Med, 6, 2014
|
|
4R4H
 
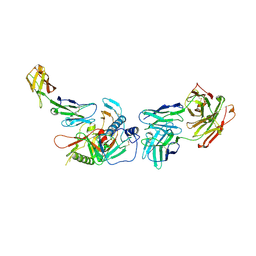 | | Crystal structure of non-neutralizing, A32-like antibody 2.2c in complex with HIV-1 Env gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c, Heavy chain, ... | | Authors: | Mclellan, J, Acharya, P, Huang, C.-C, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.28 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4R2G
 
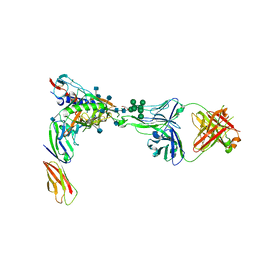 | | Crystal Structure of PGT124 Fab bound to HIV-1 JRCSF gp120 core and to CD4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Garces, F, Wilson, I.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
3S5L
 
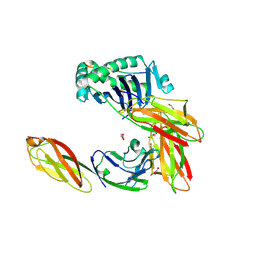 | | Crystal structure of CD4 mutant bound to HLA-DR1 | | Descriptor: | GLYCEROL, HA peptide, HLA class II histocompatibility antigen DR beta chain, ... | | Authors: | Li, Y. | | Deposit date: | 2011-05-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8QY5
 
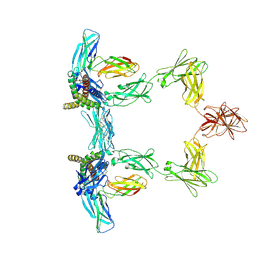 | | Structure of interleukin 6. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of interleukin 6.
To Be Published
|
|
8QY6
 
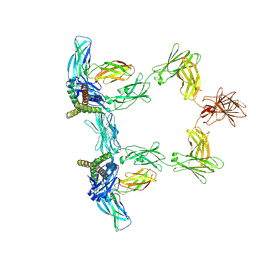 | | Structure of interleukin 6 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of interleukin 6 (gp130 P496L mutant).
To Be Published
|
|
6MEO
 
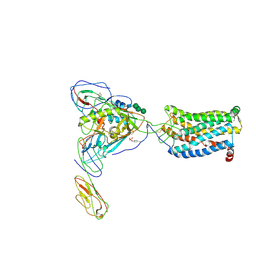 | | Structural basis of coreceptor recognition by HIV-1 envelope spike | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaik, M.M, Chen, B. | | Deposit date: | 2018-09-06 | | Release date: | 2018-12-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of coreceptor recognition by HIV-1 envelope spike.
Nature, 565, 2018
|
|
6MET
 
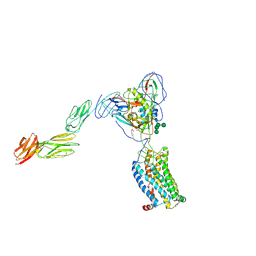 | | Structural basis of coreceptor recognition by HIV-1 envelope spike | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaik, M.M, Chen, B. | | Deposit date: | 2018-09-07 | | Release date: | 2018-12-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of coreceptor recognition by HIV-1 envelope spike.
Nature, 565, 2018
|
|
4YH7
 
 | | Crystal structure of PTPdelta ectodomain in complex with IL1RAPL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
8D82
 
 | |
2AW2
 
 | | Crystal structure of the human BTLA-HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B and T lymphocyte attenuator, NICKEL (II) ION, ... | | Authors: | Compaan, D.M, Gonzalez, L.C, Tom, I, Loyet, K.M, Eaton, D, Hymowitz, S.G. | | Deposit date: | 2005-08-31 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Attenuating Lymphocyte Activity: the crystal structure of the BTLA-HVEM complex
J.Biol.Chem., 280, 2005
|
|
2V5Y
 
 | | Crystal structure of the receptor protein tyrosine phosphatase mu ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE MU, SODIUM ION | | Authors: | Aricescu, A.R, Siebold, C, Choudhuri, K, Chang, V.T, Lu, W, Davis, S.J, van der Merwe, P.A, Jones, E.Y. | | Deposit date: | 2007-07-11 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a Tyrosine Phosphatase Adhesive Interaction Reveals a Spacer-Clamp Mechanism.
Science, 317, 2007
|
|
