6B5D
 
 | |
6PE0
 
 | | Msp1 (E214Q)-substrate complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Membrane-spanning ATPase-like protein, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6P8V
 
 | | Structure of E. coli MS115-1 HORMA:CdnC:Trip13 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, AAA family, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P12
 
 | | Structure of spastin AAA domain (wild-type) in complex with diaminotriazole-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, Drosophila melanogaster Spastin AAA domain, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94131851 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6P07
 
 | | Spastin hexamer in complex with substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sandate, C.R, Szyk, A, Zehr, E, Roll-Mecak, A, Lander, G.C. | | Deposit date: | 2019-05-16 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An allosteric network in spastin couples multiple activities required for microtubule severing.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6P14
 
 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form B) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, ACETATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93001127 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6AZ0
 
 | | Mitochondrial ATPase Protease YME1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Puchades, C, Rampello, A.J, Shin, M, Giuliano, C, Wiseman, R.L, Glynn, S.E, Lander, G.C. | | Deposit date: | 2017-09-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing.
Science, 358, 2017
|
|
6P13
 
 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form A) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, SULFATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6PDY
 
 | | Msp1-substrate complex in open conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6B5C
 
 | |
6BLB
 
 | | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP.
To be Published
|
|
6PEK
 
 | | Structure of Spastin Hexamer (Subunit A-E) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
6PDW
 
 | | Msp1-substrate complex in closed conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6PEN
 
 | | Structure of Spastin Hexamer (whole model) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, EYEYEYEYEY, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
6BMF
 
 | | Vps4p-Vta1p complex with peptide binding to the central pore of Vps4p | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Han, H, Monroe, N, Shen, P, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets.
Elife, 6, 2017
|
|
6PXI
 
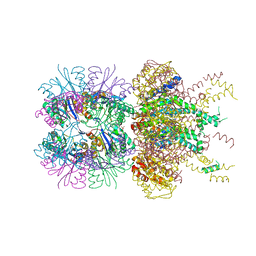 | | The crystal structure of a singly capped HslUV complex with an axial pore plug and a HslU E257Q mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.447 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
6PXK
 
 | | 3.65 Angstroms resolution structure of HslU with an axial-channel plug | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, SULFATE ION, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.647 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
6PXL
 
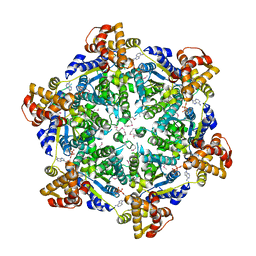 | | 3.74 Angstroms resolution structure of HlsU with an axial-channel plug | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, MAGNESIUM ION, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.741 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
6D00
 
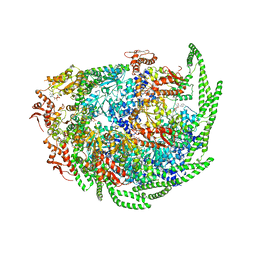 | | Calcarisporiella thermophila Hsp104 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Calcarisporiella thermophila Hsp104 | | Authors: | Zhang, K, Pintilie, G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
6QS8
 
 | |
6QS4
 
 | |
6QS7
 
 | |
6CHS
 
 | | Cdc48-Npl4 complex in the presence of ATP-gamma-S | | Descriptor: | MAGNESIUM ION, Npl4, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kim, K.H, Bodnar, N.O, Walz, T, Rapoport, T.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6QS6
 
 | |
6DJV
 
 | | Mtb ClpB in complex with ATPgammaS and casein, Conformer 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Yu, H.J, Li, H.L. | | Deposit date: | 2018-05-26 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ATP hydrolysis-coupled peptide translocation mechanism ofMycobacterium tuberculosisClpB.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
