2KXW
 
 | |
8OTW
 
 | | Cryo-EM structure of Strongylocentrotus purpuratus SLC9C1 in presence of cAMP | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sperm-specific sodium proton exchanger | | Authors: | Yeo, H, Mehta, V, Gulati, A, Drew, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure and electromechanical coupling of a voltage-gated Na + /H + exchanger.
Nature, 623, 2023
|
|
3IGA
 
 | | Potassium Channel KcsA-Fab complex in Li+ and K+ | | Descriptor: | Antibody Fab Fragment heavy chain, Antibody Fab fragment light chain, DIACYL GLYCEROL, ... | | Authors: | Thompson, A.N, Ilsoo, K, Panosian, T.D, Iverson, T.M, Allen, T.W, Nimigean, C.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3TI4
 
 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with laninamivir octanoate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-9-O-octanoyl-D-glycero-D-galacto-non-2-enonic acid, ACETATE ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TIA
 
 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
7W9K
 
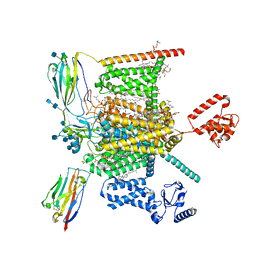 | | Cryo-EM structure of human Nav1.7-beta1-beta2 complex at 2.2 angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-09 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7XVE
 
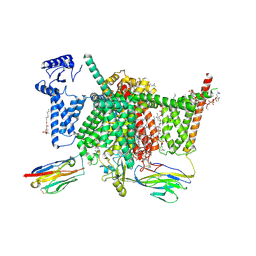 | | Human Nav1.7 mutant class-I | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XVF
 
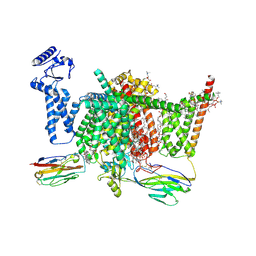 | | Nav1.7 mutant class2 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3TIC
 
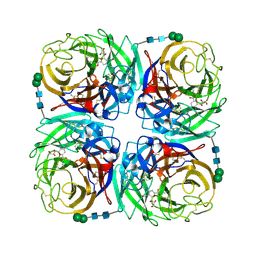 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
4OFT
 
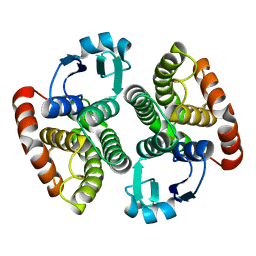 | | C- Orthorombic NaGST1 | | Descriptor: | Glutathione S-transferase-1 | | Authors: | Asojo, O.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of glutathione S-transferase 1 from the major human hookworm parasite Necator americanus (Na-GST-1) in complex with glutathione.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4OFN
 
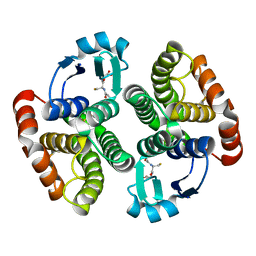 | | Monoclinic NaGST1 | | Descriptor: | GLUTATHIONE, Glutathione S-transferase-1 | | Authors: | Asojo, O.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of glutathione S-transferase 1 from the major human hookworm parasite Necator americanus (Na-GST-1) in complex with glutathione.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4HUN
 
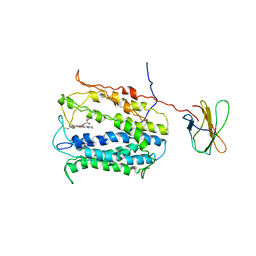 | | MATE transporter NorM-NG in complex with R6G and monobody | | Descriptor: | Multidrug efflux protein, RHODAMINE 6G, protein B | | Authors: | Lu, M. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structures of a Na+-coupled, substrate-bound MATE multidrug transporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HUL
 
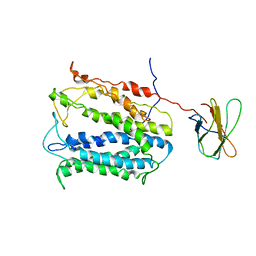 | | MATE transporter NorM-NG in complex with Cs+ and monobody | | Descriptor: | CESIUM ION, Multidrug efflux protein, Protein B | | Authors: | Lu, M. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Structures of a Na+-coupled, substrate-bound MATE multidrug transporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HUK
 
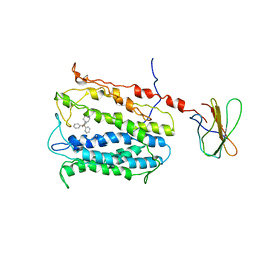 | | MATE transporter NorM-NG in complex with TPP and monobody | | Descriptor: | Multidrug efflux protein, Protein B, TETRAPHENYLPHOSPHONIUM | | Authors: | Lu, M. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structures of a Na+-coupled, substrate-bound MATE multidrug transporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HUM
 
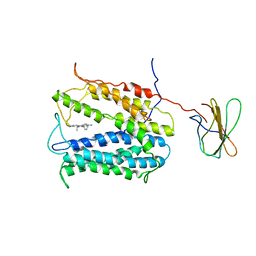 | | MATE transporter NorM-NG in complex with ethidium and monobody | | Descriptor: | ETHIDIUM, Multidrug efflux protein, Protein B | | Authors: | Lu, M. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structures of a Na+-coupled, substrate-bound MATE multidrug transporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7T9F
 
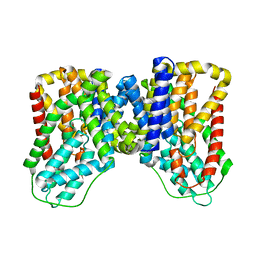 | | Structure of VcINDY-apo | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Song, J.M, Wang, D.N. | | Deposit date: | 2021-12-19 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of ion - substrate coupling in the Na + -dependent dicarboxylate transporter VcINDY.
Nat Commun, 13, 2022
|
|
4OFM
 
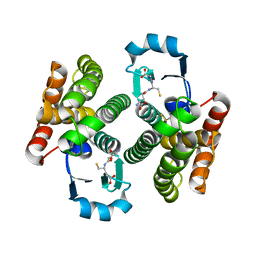 | | Triclinic NaGST1 | | Descriptor: | GLUTATHIONE, Glutathione S-transferase-1 | | Authors: | Asojo, O.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of glutathione S-transferase 1 from the major human hookworm parasite Necator americanus (Na-GST-1) in complex with glutathione.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
2MDF
 
 | |
4M8J
 
 | | Crystal structure of CaiT R262E bound to gamma-butyrobetaine | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, L-carnitine/gamma-butyrobetaine antiporter | | Authors: | Kalayil, S. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Arginine oscillation explains Na+ independence in the substrate/product antiporter CaiT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5IP5
 
 | |
5Y4O
 
 | | Cryo-EM structure of MscS channel, YnaI | | Descriptor: | Low conductance mechanosensitive channel YnaI | | Authors: | Zhang, Y, Yu, J. | | Deposit date: | 2017-08-04 | | Release date: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A binding-block ion selective mechanism revealed by a Na/K selective channel.
Protein Cell, 9, 2018
|
|
3GB7
 
 | | Potassium Channel KcsA-Fab complex in Li+ | | Descriptor: | DIACYL GLYCEROL, NICKEL (II) ION, Voltage-gated potassium channel, ... | | Authors: | Thompson, A.N, Ilsoo, K, Panosian, T.D, Iverson, T.M, Allen, T.W, Nimigean, C.M. | | Deposit date: | 2009-02-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6ZBI
 
 | | Ternary complex of Calmodulin bound to 2 molecules of NHE1 | | Descriptor: | CALCIUM ION, Calmodulin-1, Sodium/hydrogen exchanger 1 | | Authors: | Prestel, A, Kragelund, B.B, Pedersen, E.S, Pedersen, S.F, Sjoegaard-Frich, L.M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-03-17 | | Last modified: | 2021-04-14 | | Method: | SOLUTION NMR | | Cite: | Dynamic Na + /H + exchanger 1 (NHE1) - calmodulin complexes of varying stoichiometry and structure regulate Ca 2+ -dependent NHE1 activation.
Elife, 10, 2021
|
|
3E89
 
 | |
3E8G
 
 | |
