1KDX
 
 | | KIX DOMAIN OF MOUSE CBP (CREB BINDING PROTEIN) IN COMPLEX WITH PHOSPHORYLATED KINASE INDUCIBLE DOMAIN (PKID) OF RAT CREB (CYCLIC AMP RESPONSE ELEMENT BINDING PROTEIN), NMR 17 STRUCTURES | | Descriptor: | CBP, CREB | | Authors: | Radhakrishnan, I, Perez-Alvarado, G.C, Dyson, H.J, Wright, P.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-11-25 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions.
Cell(Cambridge,Mass.), 91, 1997
|
|
1U2N
 
 | | Structure CBP TAZ1 Domain | | Descriptor: | CREB binding protein, ZINC ION | | Authors: | De Guzman, R.N, Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-07-19 | | Release date: | 2005-04-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding
Biochemistry, 44, 2005
|
|
1SB0
 
 | | Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb | | Descriptor: | protein CBP, protein c-Myb | | Authors: | Zor, T, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the KIX Domain of CBP Bound to the Transactivation
Domain of c-Myb
J.Mol.Biol., 337, 2004
|
|
5I8B
 
 | | CBP in complex with Cpd23 ((R)-6-(3-(benzyloxy)phenyl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-6-[3-(benzyloxy)phenyl]-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5218 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I8G
 
 | | CBP in complex with Cpd637 ((R)-4-methyl-6-(1-methyl-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-5-yl)-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-4-methyl-6-[1-methyl-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-5-yl]-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
7R6U
 
 | | Paracoccidioides americana Pb03 Calcium Binding Protein 1 (Cbp1) | | Descriptor: | CBP domain-containing protein | | Authors: | Herrera, N, Azimova, D, Sil, A, Rosenberg, O.S. | | Deposit date: | 2021-06-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cbp1, a fungal virulence factor under positive selection, forms an effector complex that drives macrophage lysis.
Plos Pathog., 18, 2022
|
|
8C5N
 
 | | Sub-atomic resolution structure of the chitin-binding protein D (CbpD) from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Chitin-binding protein CbpD | | Authors: | Cordara, G, Krengel, U, Golten, O, Vaaje-Kolstad, G, Vinther Soerensen, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Immunization with lytic polysaccharide monooxygenase CbpD induces protective immunity against Pseudomonas aeruginosa pneumonia.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7SQX
 
 | | Crystal Structure of Pseudomonas aeruginosa lytic polysaccharide monooxygenase CbpD | | Descriptor: | AMMONIUM ION, Chitin-binding protein CbpD | | Authors: | Dade, C, Douzi, B, Ball, G, Voulhoux, R, Forest, K.T. | | Deposit date: | 2021-11-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of CbpD clarifies substrate-specificity motifs in chitin-active lytic polysaccharide monooxygenases.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WX2
 
 | | CBP-BrD complexed with NEO2734 | | Descriptor: | 1,3-dimethyl-5-[2-(oxan-4-yl)-3-[2-(trifluoromethyloxy)ethyl]benzimidazol-5-yl]pyridin-2-one, CREB-binding protein | | Authors: | Zeng, L, Lei, J.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Targeting CDCP1 gene transcription coactivated by BRD4 and CBP/p300 in castration-resistant prostate cancer.
Oncogene, 41, 2022
|
|
2KA6
 
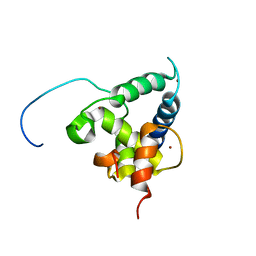 | | NMR structure of the CBP-TAZ2/STAT1-TAD complex | | Descriptor: | CREB-binding protein, Signal transducer and activator of transcription 1-alpha/beta, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Embo J., 28, 2009
|
|
1ZOQ
 
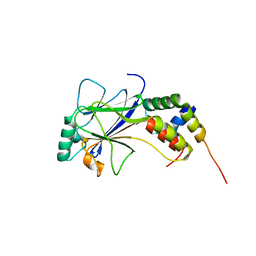 | | IRF3-CBP complex | | Descriptor: | CREB-binding protein, Interferon regulatory factor 3 | | Authors: | Qin, B, Lin, K. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of IRF-3 in complex with CBP.
Structure, 13, 2005
|
|
6KF8
 
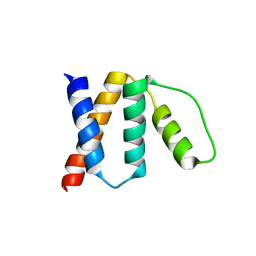 | | OsACBP2 in complex with C18:3-CoA | | Descriptor: | Acyl-CoA-binding domain-containing protein 2 | | Authors: | Jin, J, Chye, M.L. | | Deposit date: | 2019-07-06 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | OsACBP2 in complex with C18:3-CoA
To Be Published
|
|
2KQX
 
 | |
4OUF
 
 | | Crystal Structure of CBP bromodomain | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, DI(HYDROXYETHYL)ETHER | | Authors: | Roy, S, Das, C, Tyler, J.K, Kutateladze, T.G. | | Deposit date: | 2014-02-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Binding of the histone chaperone ASF1 to the CBP bromodomain promotes histone acetylation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1TOT
 
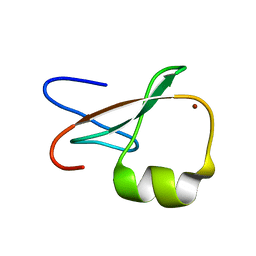 | | ZZ Domain of CBP- a Novel Fold for a Protein Interaction Module | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Legge, G.B, Martinez-Yamout, M.A, Hambly, D.M, Trinh, T, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-06-15 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module
J.Mol.Biol., 343, 2004
|
|
1F81
 
 | | SOLUTION STRUCTURE OF THE TAZ2 DOMAIN OF THE TRANSCRIPTIONAL ADAPTOR PROTEIN CBP | | Descriptor: | CREB-BINDING PROTEIN, ZINC ION | | Authors: | De Guzman, R.N, Liu, H.L, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-28 | | Release date: | 2000-10-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP.
J.Mol.Biol., 303, 2000
|
|
5XXH
 
 | | Crystal Structure Analysis of the CBP | | Descriptor: | (3S)-1-[2-(3-ethanoylindol-1-yl)ethanoyl]piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer.
Eur J Med Chem, 147, 2018
|
|
1KBH
 
 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | Descriptor: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | Authors: | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | Deposit date: | 2001-11-06 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
5NU3
 
 | | Crystal structure of the human bromodomain of CREBBP bound to the inhibitor XDM-CBP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CREB-binding protein, SULFATE ION, ... | | Authors: | Huegle, M, Wohlwend, D. | | Deposit date: | 2017-04-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2AGH
 
 | | Structural basis for cooperative transcription factor binding to the CBP coactivator | | Descriptor: | Crebbp protein, Myb proto-oncogene protein, Zinc finger protein HRX | | Authors: | De Guzman, R.N, Goto, N.K, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Cooperative Transcription Factor Binding to the CBP Coactivator
J.Mol.Biol., 355, 2006
|
|
1JSP
 
 | | NMR Structure of CBP Bromodomain in complex with p53 peptide | | Descriptor: | CREB-BINDING PROTEIN, tumor protein p53 | | Authors: | He, Y, Mujtaba, S, Zeng, L, Yan, S, Zhou, M.-M. | | Deposit date: | 2001-08-17 | | Release date: | 2002-08-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation.
Mol.Cell, 13, 2004
|
|
1JJS
 
 | | NMR Structure of IBiD, A Domain of CBP/p300 | | Descriptor: | CREB-BINDING PROTEIN | | Authors: | Lin, C.H, Hare, B.J, Wagner, G, Harrison, S.C, Maniatis, T, Fraenkel, E. | | Deposit date: | 2001-07-09 | | Release date: | 2001-10-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies.
Mol.Cell, 8, 2001
|
|
1L8C
 
 | | STRUCTURAL BASIS FOR HIF-1ALPHA/CBP RECOGNITION IN THE CELLULAR HYPOXIC RESPONSE | | Descriptor: | CREB-binding protein, Hypoxia-inducible factor 1 alpha, ZINC ION | | Authors: | Dames, S.A, Martinez-Yamout, M, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4N3W
 
 | |
4N4F
 
 | |
