4EGY
 
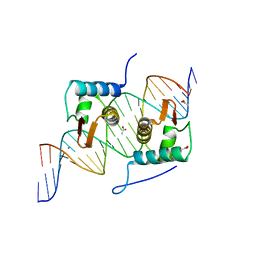 | | Crystal Structure of AraR(DBD) in complex with operator ORA1 | | Descriptor: | 5'-D(*AP*AP*AP*AP*TP*TP*GP*TP*TP*CP*GP*TP*AP*CP*AP*AP*AP*TP*AP*TP*T)-3', 5'-D(*TP*AP*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*AP*AP*CP*AP*AP*TP*TP*T)-3', ACETATE ION, ... | | Authors: | Jain, D, Nair, D.T. | | Deposit date: | 2012-04-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators.
Nucleic Acids Res., 41, 2013
|
|
4EGZ
 
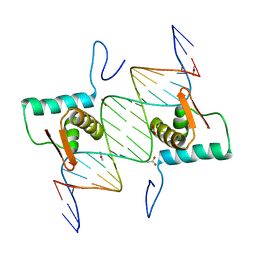 | | Crystal Structure of AraR(DBD) in complex with operator ORR3 | | Descriptor: | 5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*CP*CP*GP*TP*AP*TP*AP*CP*AP*TP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*AP*TP*GP*TP*AP*TP*AP*CP*GP*GP*AP*CP*AP*AP*AP*TP*T)-3', ACETATE ION, ... | | Authors: | Jain, D, Nair, D.T. | | Deposit date: | 2012-04-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators.
Nucleic Acids Res., 41, 2013
|
|
4H0E
 
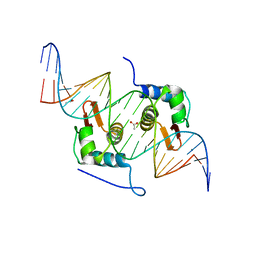 | | Crystal Structure of mutant ORR3 in complex with NTD of AraR | | Descriptor: | 5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*CP*CP*GP*TP*AP*CP*AP*TP*TP*TP*TP*AP*T)-3', 5'-D(*TP*AP*TP*AP*AP*AP*AP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*AP*AP*TP*T)-3', ACETATE ION, ... | | Authors: | Nair, D.T, Jain, D. | | Deposit date: | 2012-09-08 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators.
Nucleic Acids Res., 41, 2013
|
|
