7DP1
 
 | |
7DP0
 
 | |
7DP2
 
 | |
7X32
 
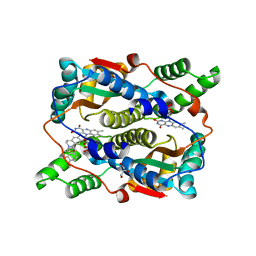 | | Crystal structure of E. coli NfsB in complex with berberine | | Descriptor: | BERBERINE, DIMETHYL SULFOXIDE, Dihydropteridine reductase, ... | | Authors: | Zhang, H, Wen, H.Y. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.829 Å) | | Cite: | Structural basis for the transformation of the traditional medicine berberine by bacterial nitroreductase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7XWW
 
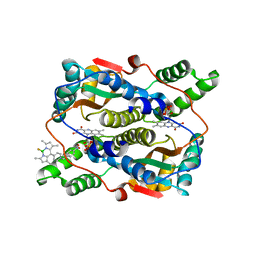 | | Crystal structure of NTR in complex with BN-XB | | Descriptor: | 2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-8-[1-[(4-nitrophenyl)methyl]pyridin-1-ium-4-yl]-3-aza-1-azonia-2-boranuidatricyclo[7.3.0.0^{3,7}]dodeca-1(12),4,6,8,10-pentaene, Dihydropteridine reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Chen, X, Chen, J, Li, J.L. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of NTR in complex with BN-XB
To Be Published
|
|
3N2S
 
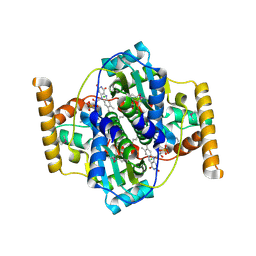 | | Structure of NfrA1 nitroreductase from B. subtilis | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NADPH-dependent nitro/flavin reductase | | Authors: | Morera, S, Gueguen-Chaignon, V, Meyer, P, Cortial, S, Ouazzani, J. | | Deposit date: | 2010-05-19 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NADH oxidase activity of Bacillus subtilis nitroreductase NfrA1: insight into its biological role.
Febs Lett., 584, 2010
|
|
4QLY
 
 | | Crystal structure of CLA-ER, a novel enone reductase catalyzing a key step of a gut-bacterial fatty acid saturation metabolism, biohydrogenation | | Descriptor: | Enone reductase CLA-ER, FLAVIN MONONUCLEOTIDE | | Authors: | Hou, F, Miyakawa, T, Tanokura, M. | | Deposit date: | 2014-06-13 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structure and reaction mechanism of a novel enone reductase.
Febs J., 282, 2015
|
|
4TTB
 
 | |
3BM2
 
 | |
3BM1
 
 | |
7Q0O
 
 | | E. coli NfsA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NADPH nitroreductase | | Authors: | White, S.A, Grainger, A, Parr, R, Day, M.A, Jarrom, D, Graziano, A, Searle, P.F, Hyde, E.I. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | The 3D-structure, kinetics and dynamics of the E. coli nitroreductase NfsA with NADP + provide glimpses of its catalytic mechanism.
Febs Lett., 596, 2022
|
|
5KRD
 
 | | Crystal structure of haliscomenobacter hydrossis iodotyrosine deiodinase (IYD) bound to FMN and 2-iodophenol (2IP) | | Descriptor: | 2-iodanylphenol, FLAVIN MONONUCLEOTIDE, Nitroreductase | | Authors: | Ingavat, N, Kavran, J.M, Sun, Z, Rokita, S. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Active Site Binding Is Not Sufficient for Reductive Deiodination by Iodotyrosine Deiodinase.
Biochemistry, 56, 2017
|
|
5KO8
 
 | | Crystal structure of haliscomenobacter hydrossis iodotyrosine deiodinase (IYD) bound to FMN and mono-iodotyrosine (I-Tyr) | | Descriptor: | 3-IODO-TYROSINE, FLAVIN MONONUCLEOTIDE, Nitroreductase | | Authors: | Ingavat, N, Kavran, J.M, Sun, Z, Rokita, S.E. | | Deposit date: | 2016-06-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Active Site Binding Is Not Sufficient for Reductive Deiodination by Iodotyrosine Deiodinase.
Biochemistry, 56, 2017
|
|
7NIY
 
 | | E. coli NfsA with FMN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-14 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NB9
 
 | | E. coli NfsA with nitrofurantoin | | Descriptor: | 1-[(~{E})-(5-nitrofuran-2-yl)methylideneamino]imidazolidine-2,4-dione, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Grainger, A.I, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NNX
 
 | | E. coli NfsA with 1,4-benzoquinone | | Descriptor: | 1,2-ETHANEDIOL, 1,4-benzoquinone, CHLORIDE ION, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NMP
 
 | | E. coli NfsA with hydroquinone | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Day, M.D, Jarrom, D, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
5KO7
 
 | | Crystal structure of haliscomenobacter hydrossis iodotyrosine deiodinase (IYD) bound to FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase | | Authors: | Ingavat, N, Kavran, J.M, Sun, Z, Rokita, S. | | Deposit date: | 2016-06-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Active Site Binding Is Not Sufficient for Reductive Deiodination by Iodotyrosine Deiodinase.
Biochemistry, 56, 2017
|
|
7RZP
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2866 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Dihydropteridine reductase, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-27 | | Release date: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2866
To Be Published
|
|
7S1A
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae Rd KW20 | | Descriptor: | ACETIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-01 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae Rd KW20
To Be Published
|
|
7S14
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae 86-028NP | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-31 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae 86-028NP
To Be Published
|
|
7T2Z
 
 | |
7T33
 
 | |
8CQS
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_0217) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase-like protein, PHOSPHATE ION | | Authors: | Blaha, J, Gratzl, S, Mortensen, S.A, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQU
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_1680) | | Descriptor: | CITRIC ACID, Putative NADH dehydrogenase/NAD(P)H nitroreductase, TERTIARY-BUTYL ALCOHOL | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
