7JPS
 
 | |
7K59
 
 | | Structure of apo VCP hexamer generated from bacterially recombinant VCP/p97 | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
7K57
 
 | | Structure of apo VCP dodecamer generated from bacterially recombinant VCP/p97 | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
7K56
 
 | | Structure of VCP dodecamer purified from H1299 cells | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
7KSL
 
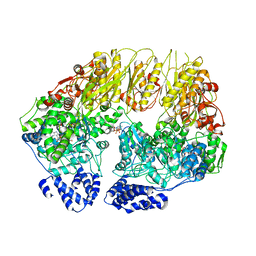 | | Substrate-free human mitochondrial LONP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7KSM
 
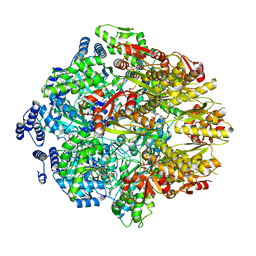 | | Human mitochondrial LONP1 with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7KRZ
 
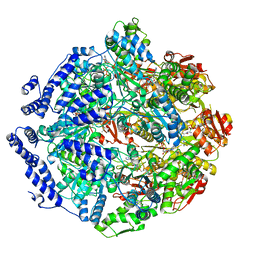 | | Human mitochondrial LONP1 in complex with Bortezomib | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Endogenous co-purified substrate, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-20 | | Release date: | 2021-02-24 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7L9P
 
 | | Structure of human SHLD2-SHLD3-REV7-TRIP13(E253Q) complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pachytene checkpoint protein 2 homolog, ... | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2021-01-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L9X
 
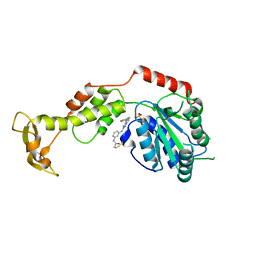 | | Structure of VPS4B in complex with an allele-specific covalent inhibitor | | Descriptor: | N-{3-[(8-phenyl[1,2,4]triazolo[1,5-a]pyridin-2-yl)amino]phenyl}propanamide, SULFATE ION, Vacuolar protein sorting-associated protein 4B | | Authors: | Grasso, M, Cupido, T, Kapoor, T.M. | | Deposit date: | 2021-01-05 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A chemical genetics approach to examine the functions of AAA proteins.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L6N
 
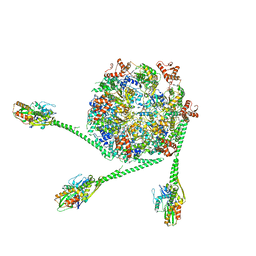 | | The Mycobacterium tuberculosis ClpB disaggregase hexamer structure with three locally refined ClpB middle domains and three DnaK nucleotide binding domains | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB, Chaperone protein DnaK, ... | | Authors: | Yin, Y.Y, Feng, X, Li, H. | | Deposit date: | 2020-12-23 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis for aggregate dissolution and refolding by the Mycobacterium tuberculosis ClpB-DnaK bi-chaperone system.
Cell Rep, 35, 2021
|
|
7L5W
 
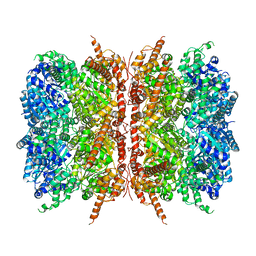 | | p97-R155H mutant dodecamer I | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7L5X
 
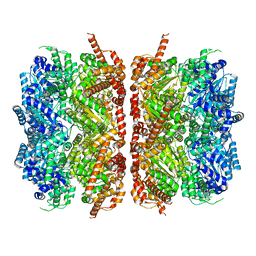 | | p97-R155H mutant dodecamer II | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7LJF
 
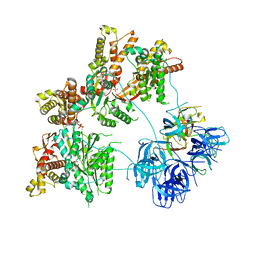 | |
7LN5
 
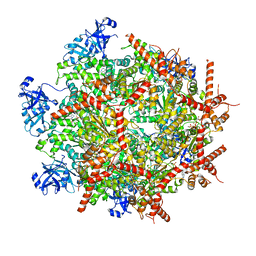 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (CHAPSO, Class 1, Close State) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN4
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN0
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LMY
 
 | | Cryo-EM structure of human p97 in complex with NMS-873 in the presence of ATP, Npl4/Ufd1, and Ub6 | | Descriptor: | 3-[3-cyclopentylsulfanyl-5-[[3-methyl-4-(4-methylsulfonylphenyl)phenoxy]methyl]-1,2,4-triazol-4-yl]pyridine, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN2
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN3
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LMZ
 
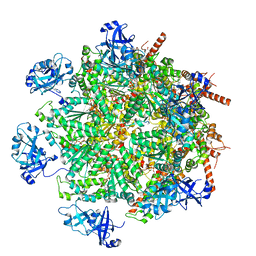 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN1
 
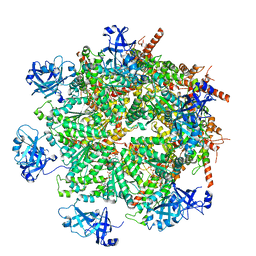 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN6
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (CHAPSO, Class 2, Open State) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MHB
 
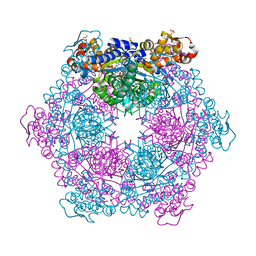 | |
7MCA
 
 | |
7MDM
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
