6W6G
 
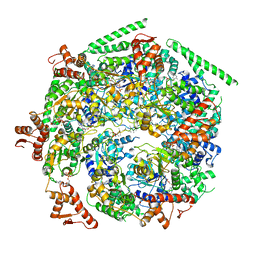 | |
6W6H
 
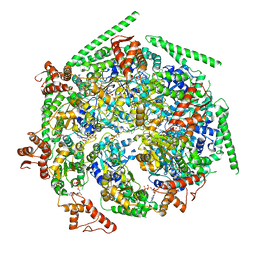 | |
6W6I
 
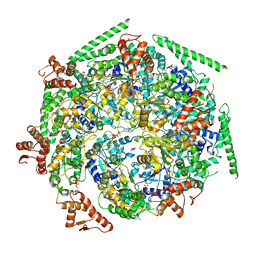 | |
6W6E
 
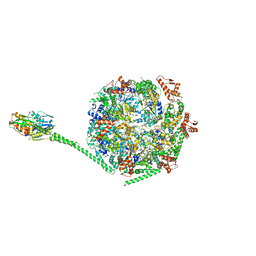 | |
6W6J
 
 | |
6WGI
 
 | | Atomic model of the mutant OCCM (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) loaded on DNA at 10.5 A resolution | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (34-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WJD
 
 | |
6WGG
 
 | | Atomic model of pre-insertion mutant OCCM-DNA complex(ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (41-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGC
 
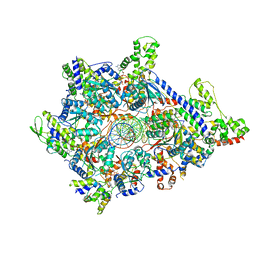 | | Atomic model of semi-attached mutant OCCM-DNA complex (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, DNA (41-MER), DNA replication licensing factor MCM3, ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WJN
 
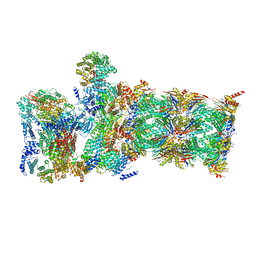 | |
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | Authors: | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | Deposit date: | 2020-04-28 | | Release date: | 2021-06-09 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
7X7Q
 
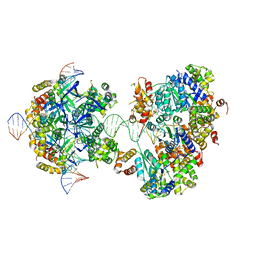 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | Descriptor: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X5B
 
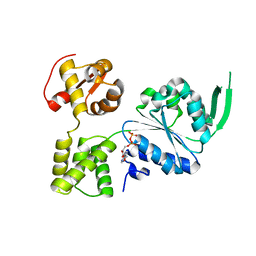 | | Crystal structure of RuvB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z, Dai, L. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7P
 
 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | Descriptor: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7YKK
 
 | | Cryo-EM structure of Drg1 hexamer treated with ADP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKT
 
 | | Cryo-EM structure of Drg1 hexamer in helical state treated with ADP/AMPPNP/benzo-diazaborine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-23 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKZ
 
 | | Cryo-EM structure of Drg1 hexamer in the planar state treated with ADP/AMPPNP/Diazaborine | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKL
 
 | | Cryo-EM structure of Drg1 hexamer treated with AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7Y59
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y53
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Z6H
 
 | | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17, Checkpoint protein HUS1, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Nucleic Acids Res., 50, 2022
|
|
7Z11
 
 | | Structure of substrate bound DRG1 (AFG2) | | Descriptor: | ATPase family gene 2 protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, peptide substrate | | Authors: | Prattes, M, Grishkovskaya, I, Bergler, H, Haselbach, D. | | Deposit date: | 2022-02-24 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z34
 
 | | Structure of pre-60S particle bound to DRG1(AFG2). | | Descriptor: | 35S pre-ribosomal RNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Prattes, M, Grishkovskaya, I, Bergler, H, Haselbach, D. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-21 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZBH
 
 | |
