1EQT
 
 | | MET-RANTES | | Descriptor: | SULFATE ION, T-CELL SPECIFIC RANTES PROTEIN | | Authors: | Hoover, D.M, Shaw, J, Gryczynski, Z, Proudfoot, A.E.I, Wells, T. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of MET-RANTES: Comparison with Native RANTES and AOP-RANTES
PROTEIN PEPT.LETT., 7, 2000
|
|
1EIH
 
 | |
1EL0
 
 | | SOLUTION STRUCTURE OF THE HUMAN CC CHEMOKINE, I-309 | | Descriptor: | I-309 | | Authors: | Keizer, D.W, Crump, M.P, Lee, T.W, Slupsky, C.M, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-03-11 | | Release date: | 2000-09-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Human CC chemokine I-309, structural consequences of the additional disulfide bond.
Biochemistry, 39, 2000
|
|
2EOT
 
 | | SOLUTION STRUCTURE OF EOTAXIN, AN ENSEMBLE OF 32 NMR SOLUTION STRUCTURES | | Descriptor: | EOTAXIN | | Authors: | Crump, M.P, Rajarathnam, K, Kim, K.-S, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1998-06-29 | | Release date: | 1998-11-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of eotaxin, a chemokine that selectively recruits eosinophils in allergic inflammation.
J.Biol.Chem., 273, 1998
|
|
2FJ2
 
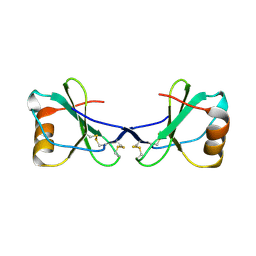 | | Crystal Structure of Viral Macrophage Inflammatory Protein-II | | Descriptor: | Viral macrophage inflammatory protein-II | | Authors: | Li, Y, Liu, D, Cao, R, Kumar, S, Dong, C.Z, wilson, S.R, Gao, Y.G, Huang, Z. | | Deposit date: | 2005-12-30 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of chemically synthesized vMIP-II.
Proteins, 67, 2007
|
|
2FHT
 
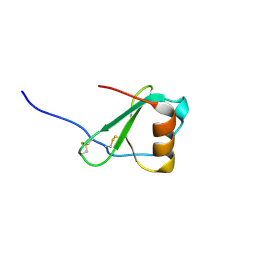 | | Crystal Structure of Viral Macrophage Inflammatory Protein-II | | Descriptor: | Viral macrophage inflammatory protein-II | | Authors: | Li, Y, Liu, D, Cao, R, Kumar, S, Dong, C.Z, wilson, S.R, Gao, Y.G, Huang, Z. | | Deposit date: | 2005-12-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chemically synthesized vMIP-II.
Proteins, 67, 2007
|
|
2HCI
 
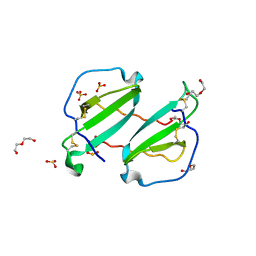 | | Structure of Human Mip-3a Chemokine | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, SULFITE ION, ... | | Authors: | Malik, Z.A, Tack, B.F. | | Deposit date: | 2006-06-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of human MIP-3alpha chemokine.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2HDL
 
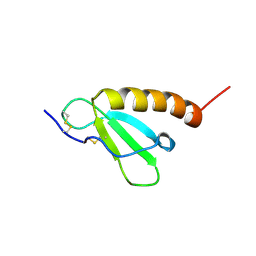 | | Solution structure of Brak/CXCL14 | | Descriptor: | Small inducible cytokine B14 | | Authors: | Peterson, F.C, Thorpe, J.A, Harder, A.G, Volkman, B.F, Schwarze, S.R. | | Deposit date: | 2006-06-20 | | Release date: | 2006-10-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Determinants Involved in the Regulation of CXCL14/BRAK Expression by the 26 S Proteasome.
J.Mol.Biol., 363, 2006
|
|
2HDM
 
 | |
2HCC
 
 | | SOLUTION STRUCTURE OF THE HUMAN CHEMOKINE HCC-2, NMR, 30 STRUCTURES | | Descriptor: | HUMAN CHEMOKINE HCC-2 | | Authors: | Sticht, H, Escher, S.E, Schweimer, K, Forssmann, W.G, Roesch, P, Adermann, K. | | Deposit date: | 1998-07-03 | | Release date: | 1999-07-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human CC chemokine 2: A monomeric representative of the CC chemokine subtype.
Biochemistry, 38, 1999
|
|
4ZLT
 
 | |
5COY
 
 | | Crystal structure of CC chemokine 5 (CCL5) | | Descriptor: | C-C motif chemokine 5, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-07-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CMD
 
 | |
5COR
 
 | |
5D14
 
 | |
5D65
 
 | | X-RAY STRUCTURE OF MACROPHAGE INFLAMMATORY PROTEIN-1 ALPHA (CCL3) WITH HEPARIN COMPLEX | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-C motif chemokine 3, CHLORIDE ION, ... | | Authors: | Liang, W.G, Hwang, D.Y, Zulueta, M.M, Hung, S.C, Tang, W. | | Deposit date: | 2015-08-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DNF
 
 | | Crystal structure of CC chemokine 5 (CCL5) oligomer in complex with heparin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, C-C motif chemokine 5, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1JE4
 
 | |
1LV9
 
 | | CXCR3 Binding Chemokine IP-10/CXCL10 | | Descriptor: | Small inducible cytokine B10 | | Authors: | Booth, V, Keizer, D.W, Kamphuis, M.B, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2002-05-27 | | Release date: | 2002-09-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The CXCR3 binding chemokine IP-10/CXCL10: structure and receptor interactions.
Biochemistry, 41, 2002
|
|
1M8A
 
 | | Human MIP-3alpha/CCL20 | | Descriptor: | ISOPROPYL ALCOHOL, Small inducible cytokine A20 | | Authors: | Hoover, D.M, Boulegue, C, Yang, D, Oppenheim, J.J, Tucker, K, Lu, W, Lubkowski, J. | | Deposit date: | 2002-07-24 | | Release date: | 2002-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of human macrophage inflammatory protein-3alpha /CCL20. Linking antimicrobial and CC chemokine receptor-6-binding activities with human beta-defensins
J.Biol.Chem., 277, 2002
|
|
1MI2
 
 | | SOLUTION STRUCTURE OF MURINE MACROPHAGE INFLAMMATORY PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN-2 | | Authors: | Shao, W, Jerva, L.F, West, J, Lolis, E, Schweitzer, B.I. | | Deposit date: | 1997-10-24 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of murine macrophage inflammatory protein-2.
Biochemistry, 37, 1998
|
|
1MGS
 
 | |
1MSH
 
 | |
1NAP
 
 | |
1MSG
 
 | |
