6YFQ
 
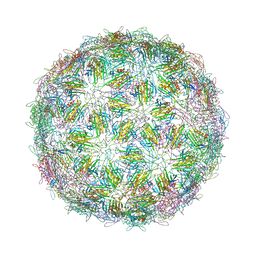 | | Virus-like particle of bacteriophage NT-214 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YJI
 
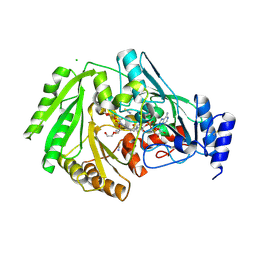 | | Structure of FgCelDH7C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Haddad Momeni, M, Fredslund, F, Berrin, J.G, Abou Hachem, M, Welner, D.H. | | Deposit date: | 2020-04-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of fungal oligosaccharide-oxidising flavo-enzymes with previously unknown substrates, redox-activity profiles and interplay with LPMOs.
Nat Commun, 12, 2021
|
|
5E13
 
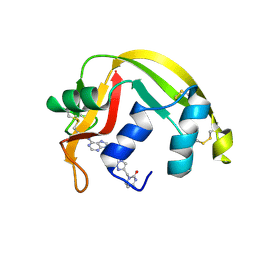 | | Crystal structure of Eosinophil-derived neurotoxin in complex with the triazole double-headed ribonucleoside 11c | | Descriptor: | 3'-{4-[(4-amino-2-oxopyrimidin-1(2H)-yl)methyl]-1H-1,2,3-triazol-1-yl}-3'-deoxyadenosine, Non-secretory ribonuclease | | Authors: | Chatzileontiadou, D.S.M, Stravodimos, G.A, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Triazole double-headed ribonucleosides as inhibitors of eosinophil derived neurotoxin.
Bioorg.Chem., 63, 2015
|
|
5ED0
 
 | |
5DWC
 
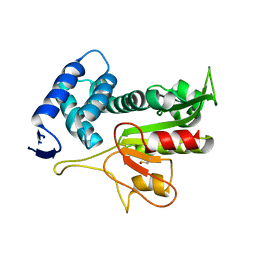 | |
5ECY
 
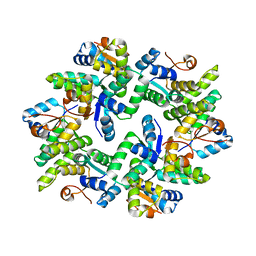 | |
6ZCT
 
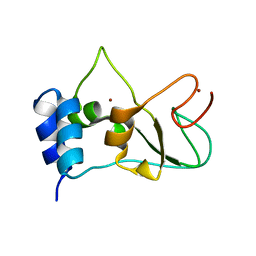 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | ZINC ION, nsp10 | | Authors: | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
5EHB
 
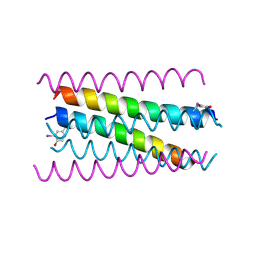 | | A de novo designed hexameric coiled-coil peptide with iodotyrosine | | Descriptor: | pHiosYI | | Authors: | Lizatovic, R, Aurelius, O, Stenstrom, O, Drakenberg, T, Akke, M, Logan, D.T, Andre, I. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-15 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | A De Novo Designed Coiled-Coil Peptide with a Reversible pH-Induced Oligomerization Switch.
Structure, 24, 2016
|
|
6ZLF
 
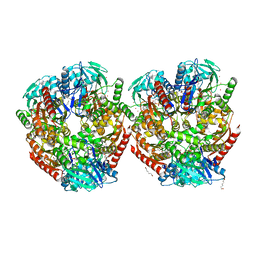 | | Aerobic crystal structure of F420H2-Oxidase from Methanothermococcus thermolithotrophicus at 1.8A resolution under 125 bars of krypton | | Descriptor: | CHLORIDE ION, Coenzyme F420H2 oxidase (FprA), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Engilberge, S, Wagner, T, Carpentier, P, Girard, E, Shima, S. | | Deposit date: | 2020-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Krypton-derivatization highlights O 2 -channeling in a four-electron reducing oxidase.
Chem.Commun.(Camb.), 56, 2020
|
|
5EOP
 
 | |
6YUH
 
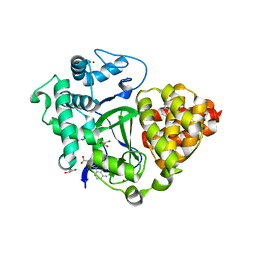 | | Crystal structure of SMYD3 with diperodon R enantiomer bound to allosteric site | | Descriptor: | Diperodon, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Cederfelt, D, Talibov, V.O, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of an Allosteric Ligand Binding Site in SMYD3 Lysine Methyltransferase.
Chembiochem, 22, 2021
|
|
6ZB8
 
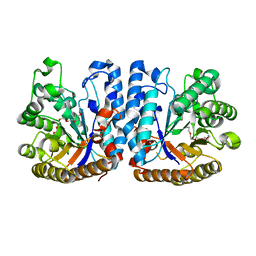 | | Exo-beta-1,3-glucanase from moose rumen microbiome, active site mutant E167Q/E295Q | | Descriptor: | Exo-beta-1,3-glucanase variant E167Q/E295Q, POLYETHYLENE GLYCOL (N=34) | | Authors: | Kalyani, D.C, Reichenbach, T, Aspeborg, H, Divne, C. | | Deposit date: | 2020-06-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A homodimeric bacterial exo-beta-1,3-glucanase derived from moose rumen microbiome shows a structural framework similar to yeast exo-beta-1,3-glucanases.
Enzyme.Microb.Technol., 143, 2021
|
|
5EL2
 
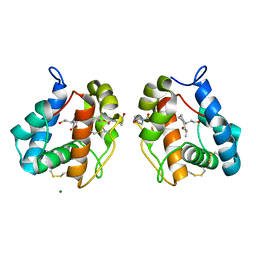 | |
6ZK8
 
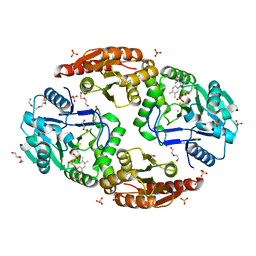 | | Native crystal structure of anaerobic F420H2-Oxidase from Methanothermococcus thermolithotrophicus at 1.8A resolution | | Descriptor: | Coenzyme F420H2 oxidase (FprA), DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Engilberge, S, Wagner, T, Carpentier, P, Girard, E, Shima, S. | | Deposit date: | 2020-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Krypton-derivatization highlights O 2 -channeling in a four-electron reducing oxidase.
Chem.Commun.(Camb.), 56, 2020
|
|
6Z7W
 
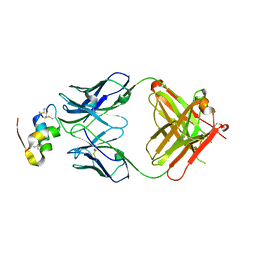 | | Human insulin in complex with the analytical antibody HUI-018 Fab | | Descriptor: | HUI-018 Fab Heavy Chain, Insulin, MAb 6H10 light chain | | Authors: | Johansson, E. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Insulin binding to the analytical antibody sandwich pair OXI-005 and HUI-018: Epitope mapping and binding properties.
Protein Sci., 30, 2021
|
|
6ZF8
 
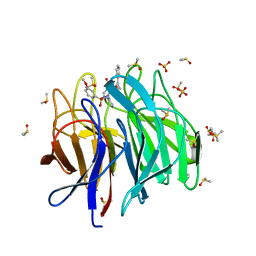 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[2-hydroxy-2-oxoethyl-(3-methoxyphenyl)sulfonyl-amino]phenyl]-5-[(1~{S},2~{S})-2-phenylcyclopropyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6YXK
 
 | | Crystal structure of ACPA 3F3 in complex with cit-vimentin 59-74 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACPA 3F3 Fab fragment - heavy chain, ACPA 3F3 Fab fragment - light chain, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Surface Ig variable domain glycosylation affects autoantigen binding and acts as threshold for human autoreactive B cell activation.
Sci Adv, 8, 2022
|
|
6YXL
 
 | | Crystal structure of ACPA F3 | | Descriptor: | ACPA F3 Fab fragment - heavy chain, ACPA F3 Fab fragment - light chain, GLYCEROL, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Surface Ig variable domain glycosylation affects autoantigen binding and acts as threshold for human autoreactive B cell activation.
Sci Adv, 8, 2022
|
|
6Z2T
 
 | |
5ETZ
 
 | |
5ECW
 
 | |
5ELV
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L504-N775) in complex with glutamate and BPAM-521 at 1.92 A resolution | | Descriptor: | 4-Cyclopropyl-3,4-dihydro-7-hydroxy-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Krintel, C, Juknaite, L, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-11-05 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Enthalpy-Entropy Compensation in the Binding of Modulators at Ionotropic Glutamate Receptor GluA2.
Biophys.J., 110, 2016
|
|
6XYK
 
 | |
6ZPE
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Replicase polyprotein 1ab, ... | | Authors: | Fisher, S.Z, Kozielski, F. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
5E94
 
 | |
