6L3P
 
 | |
6LDZ
 
 | | Crystal structure of Rv0222 from Mycobacterium tuberculosis | | Descriptor: | Probable enoyl-CoA hydratase EchA1 (Enoyl hydrase) (Unsaturated acyl-CoA hydratase) (Crotonase) | | Authors: | Li, J, Ran, Y.J, Wang, L, Wu, J.H, Ge, B.X, Rao, Z.H. | | Deposit date: | 2019-11-23 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Host-mediated ubiquitination of a mycobacterial protein suppresses immunity.
Nature, 577, 2020
|
|
4U1A
 
 | | Crystal structure of human peroxisomal delta3,delta2, enoyl-CoA isomerase helix-10 deletion mutant (ISOB-ECI2) | | Descriptor: | CHLORIDE ION, Enoyl-CoA delta isomerase 2 | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Human Delta (3) , Delta (2) -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10.
Febs J., 282, 2015
|
|
4U19
 
 | | Crystal structure of human peroxisomal delta3,delta2, enoyl-CoA isomerase V349A mutant (ISOA-ECI2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enoyl-CoA delta isomerase 2 | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Human Delta (3) , Delta (2) -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10.
Febs J., 282, 2015
|
|
8SK6
 
 | | human liver mitochondrial Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase | | Descriptor: | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | | Authors: | Zhang, Z, Tringides, M. | | Deposit date: | 2023-04-18 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
4U18
 
 | | Crystal structure of human peroxisomal delta3,delta2, enoyl-CoA isomerase (ISO-ECI2) | | Descriptor: | CHLORIDE ION, Enoyl-CoA delta isomerase 2, mitochondrial, ... | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Human Delta (3) , Delta (2) -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10.
Febs J., 282, 2015
|
|
1K39
 
 | | The structure of yeast delta3-delta2-enoyl-COA isomerase complexed with octanoyl-COA | | Descriptor: | OCTANOYL-COENZYME A, PHOSPHATE ION, d3,d2-enoyl CoA isomerase ECI1 | | Authors: | Mursula, A.M, Geerlof, A, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: |
|
|
8SOY
 
 | |
8UJA
 
 | |
1HNU
 
 | | CRYSTAL STRUCTURE OF PEROXISOMAL DELTA3-DELTA2-ENOYL-COA ISOMERASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 1,2-ETHANEDIOL, D3,D2-ENOYL COA ISOMERASE ECI1, PERRHENATE | | Authors: | Mursula, A.M, van Aalten, D.M.F, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2000-12-08 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of delta(3)-delta(2)-enoyl-CoA isomerase.
J.Mol.Biol., 309, 2001
|
|
1HNO
 
 | | CRYSTAL STRUCTURE OF PEROXISOMAL DELTA3-DELTA2-ENOYL-COA ISOMERASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 1,2-ETHANEDIOL, D3,D2-ENOYL COA ISOMERASE ECI1 | | Authors: | Mursula, A.M, van Aalten, D.M.F, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2000-12-08 | | Release date: | 2001-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of delta(3)-delta(2)-enoyl-CoA isomerase.
J.Mol.Biol., 309, 2001
|
|
1HZD
 
 | | CRYSTAL STRUCTURE OF HUMAN AUH PROTEIN, AN RNA-BINDING HOMOLOGUE OF ENOYL-COA HYDRATASE | | Descriptor: | AU-BINDING PROTEIN/ENOYL-COA HYDRATASE | | Authors: | Kurimoto, K, Fukai, S, Nureki, O, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-01-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human AUH protein, a single-stranded RNA binding homolog of enoyl-CoA hydratase.
Structure, 9, 2001
|
|
7XWC
 
 | |
7XWT
 
 | |
7XWV
 
 | |
1JXZ
 
 | | Structure of the H90Q mutant of 4-Chlorobenzoyl-Coenzyme A Dehalogenase complexed with 4-hydroxybenzoyl-Coenzyme A (product) | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-chlorobenzoyl Coenzyme A dehalogenase, CALCIUM ION, ... | | Authors: | Thoden, J.B, Zhang, W, Wei, Y, Luo, L, Taylor, K.L, Yang, G, Dunaway-Mariano, D, Benning, M.M, Holden, H.M. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 90 Function in 4-chlorobenzoyl-coenzyme A Dehalogenase Catalysis
Biochemistry, 40, 2001
|
|
1MJ3
 
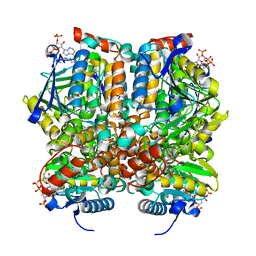 | | Crystal Structure Analysis of rat enoyl-CoA hydratase in complex with hexadienoyl-CoA | | Descriptor: | ENOYL-COA HYDRATASE, MITOCHONDRIAL, HEXANOYL-COENZYME A | | Authors: | Bell, A.F, Feng, Y, Hofstein, H.A, Parikh, S, Wu, J, Rudolph, M.J, Kisker, C, Tonge, P.J. | | Deposit date: | 2002-08-26 | | Release date: | 2002-09-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stereoselectivity of Enoyl-CoA Hydratase Results from Preferential Activation of
One of Two Bound Substrate Conformers
Chem.Biol., 9, 2002
|
|
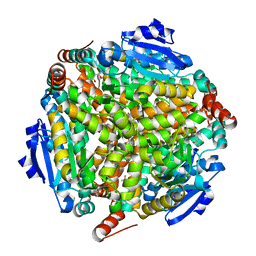
1WZ8
 
 | |
5KAJ
 
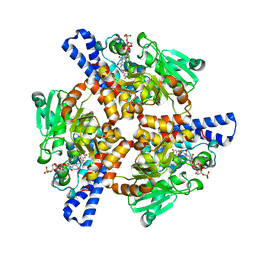 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21, A319C mutant | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[(~{E})-2-[3,5-bis(oxidanyl)phenyl]-1-oxidanyl-ethenyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1XX4
 
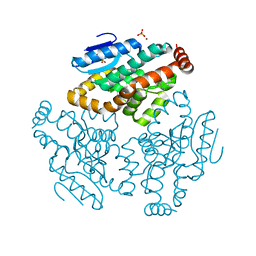 | | Crystal Structure of Rat Mitochondrial 3,2-Enoyl-CoA | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, mitochondrial, BENZAMIDINE, ... | | Authors: | Hubbard, P.A, Yu, W, Schulz, H, Kim, J.-J. | | Deposit date: | 2004-11-03 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain swapping in the low-similarity isomerase/hydratase superfamily: the crystal structure of rat mitochondrial Delta3, Delta2-enoyl-CoA isomerase.
Protein Sci., 14, 2005
|
|
5KAH
 
 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21, V425T mutant | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.779 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5KAG
 
 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21 | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, OXYGEN MOLECULE, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6SLB
 
 | | Crystal structure of isomerase PaaG with trans-3,4-didehydroadipyl-CoA | | Descriptor: | (~{E})-6-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]-6-oxidanylidene-hex-3-enoic acid, Enoyl-CoA hydratase/carnithine racemase | | Authors: | Saleem-Batcha, R, Spieker, M, Teufel, R. | | Deposit date: | 2019-08-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and Mechanistic Basis of an Oxepin-CoA Forming Isomerase in Bacterial Primary and Secondary Metabolism.
Acs Chem.Biol., 14, 2019
|
|
5KJP
 
 | |
6SLA
 
 | | Crystal structure of isomerase PaaG mutant - D136N with Oxepin-CoA | | Descriptor: | Enoyl-CoA hydratase/carnithine racemase, ~{S}-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 2-(2,5-dihydrooxepin-7-yl)ethanethioate | | Authors: | Saleem-Batcha, R, Spieker, M, Teufel, R. | | Deposit date: | 2019-08-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Basis of an Oxepin-CoA Forming Isomerase in Bacterial Primary and Secondary Metabolism.
Acs Chem.Biol., 14, 2019
|
|
