3EHQ
 
 | | Crystal Structure of Human Osteoclast Stimulating Factor | | Descriptor: | 1,2-ETHANEDIOL, Osteoclast-stimulating factor 1 | | Authors: | Tong, S, Zhou, H, Gao, Y, Zhu, Z, Zhang, X, Teng, M, Niu, L. | | Deposit date: | 2008-09-14 | | Release date: | 2009-08-04 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of human osteoclast stimulating factor
Proteins, 75, 2009
|
|
3UAT
 
 | |
4A63
 
 | | Crystal structure of the p73-ASPP2 complex at 2.6A resolution | | Descriptor: | ACETATE ION, APOPTOSIS STIMULATING OF P53 PROTEIN 2, TUMOUR PROTEIN 73, ... | | Authors: | Canning, P, Sharpe, T, Krojer, T, Savitsky, P, Cooper, C.D.O, Salah, E, Keates, T, Muniz, J, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C, Bountra, C, Edwards, A, Bullock, A.N. | | Deposit date: | 2011-10-31 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis for Aspp2 Recognition by the Tumor Suppressor P73.
J.Mol.Biol., 423, 2012
|
|
5NUH
 
 | |
5FWA
 
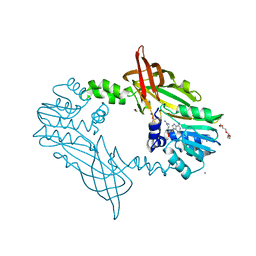 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP1 | | Descriptor: | 1,2-ETHANEDIOL, 9-(6-carbamimidamido-5,6-dideoxy-beta-D-ribo-hexofuranosyl)-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5FUL
 
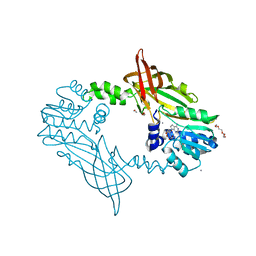 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with SAH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5FWD
 
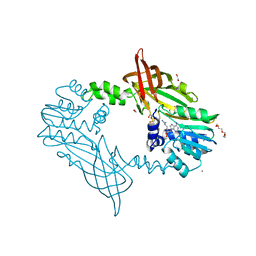 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP2 | | Descriptor: | 1,2-ETHANEDIOL, 9-(7-{[amino(iminio)methyl]amino}-5,6,7-trideoxy-beta-D-ribo-heptofuranosyl)-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-12 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies of Protein Arginine Methyltransferase 2 Reveal its Interactions with Potential Substrates and Inhibitors.
FEBS J., 284, 2017
|
|
2ROL
 
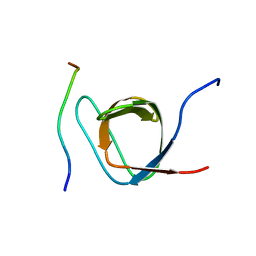 | | Structural Basis of PxxDY motif recognition in SH3 binding | | Descriptor: | 12-meric peptide from T-cell surface glycoprotein CD3 epsilon chain, Epidermal growth factor receptor kinase substrate 8-like protein 1 | | Authors: | Aitio, O, Hellman, M, Kesti, T, Kleino, I, Samuilova, O, Tossavainen, H, Paakkonen, K, Saksela, K, Permi, P. | | Deposit date: | 2008-04-02 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of PxxDY motif recognition in SH3 binding
J.Mol.Biol., 382, 2008
|
|
2LZ6
 
 | | Distinct ubiquitin binding modes exhibited by sh3 domains: molecular determinants and functional implications | | Descriptor: | CD2-associated protein, Ubiquitin | | Authors: | Ortega-Roldan, J, Azuaga, A, Blackledge, M, Van Nuland, N. | | Deposit date: | 2012-09-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distinct Ubiquitin Binding Modes Exhibited by SH3 Domains: Molecular Determinants and Functional Implications.
Plos One, 8, 2013
|
|
2MOX
 
 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2014-05-07 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
2PZ1
 
 | | Crystal Structure of Auto-inhibited Asef | | Descriptor: | Rho guanine nucleotide exchange factor 4 | | Authors: | Betts, L, Sondek, J, Rossman, K.L. | | Deposit date: | 2007-05-17 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Release of autoinhibition of ASEF by APC leads to CDC42 activation and tumor suppression.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1NG2
 
 | | Structure of autoinhibited p47phox | | Descriptor: | Neutrophil cytosolic factor 1 | | Authors: | Groemping, Y, Lapouge, K, Smerdon, S.J, Rittinger, K. | | Deposit date: | 2002-12-16 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of phosphorylation-induced activation of the NADPH oxidase
Cell(Cambridge,Mass.), 113, 2003
|
|
1JXO
 
 | |
1K4U
 
 | | Solution structure of the C-terminal SH3 domain of p67phox complexed with the C-terminal tail region of p47phox | | Descriptor: | PHAGOCYTE NADPH OXIDASE SUBUNIT P47PHOX, PHAGOCYTE NADPH OXIDASE SUBUNIT P67PHOX | | Authors: | Kami, K, Takeya, R, Sumimoto, H, Kohda, D. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p.
EMBO J., 21, 2002
|
|
1JXM
 
 | | CRYSTAL STRUCTURE OF THE GMP BOUND SH3-HOOK-GK FRAGMENT OF PSD-95 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANIDINE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Tavares, G.A, Panepucci, E.H, Brunger, A.T. | | Deposit date: | 2001-09-07 | | Release date: | 2002-01-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the intramolecular interaction between the SH3 and guanylate kinase domains of PSD-95.
Mol.Cell, 8, 2001
|
|
1KJW
 
 | | SH3-Guanylate Kinase Module from PSD-95 | | Descriptor: | POSTSYNAPTIC DENSITY PROTEIN 95, SULFATE ION | | Authors: | McGee, A.W, Dakoji, S.R, Olsen, O, Bredt, D.S, Lim, W.A, Prehoda, K.E. | | Deposit date: | 2001-12-05 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the SH3-Guanylate Kinase Module from PSD-95 Suggests a Mechanism for Regulated Assembly of MAGUK Scaffolding Proteins
Mol.Cell, 8, 2001
|
|
1U5S
 
 | | NMR structure of the complex between Nck-2 SH3 domain and PINCH-1 LIM4 domain | | Descriptor: | Cytoplasmic protein NCK2, PINCH protein, ZINC ION | | Authors: | Vaynberg, J, Fukuda, T, Vinogradova, O, Velyvis, A, Ng, L, Wu, C, Qin, J. | | Deposit date: | 2004-07-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility.
Mol.Cell, 17, 2005
|
|
1UEC
 
 | | Crystal structure of autoinhibited form of tandem SH3 domain of p47phox | | Descriptor: | Neutrophil cytosol factor 1 | | Authors: | Yuzawa, S, Suzuki, N.N, Fujioka, Y, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2003-05-11 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A molecular mechanism for autoinhibition of the tandem SH3 domains of p47phox, the regulatory subunit of the phagocyte NADPH oxidase
Genes Cells, 9, 2004
|
|
1AVZ
 
 | |
1EFN
 
 | |
1YCS
 
 | | P53-53BP2 COMPLEX | | Descriptor: | 53BP2, P53, ZINC ION | | Authors: | Gorina, S, Pavletich, N.P. | | Deposit date: | 1996-09-30 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2.
Science, 274, 1996
|
|
2JT4
 
 | |
2KBT
 
 | | Attachment of an NMR-invisible solubility enhancement tag (INSET) using a sortase-mediated protein ligation method | | Descriptor: | Proto-oncogene vav,Immunoglobulin G-binding protein G | | Authors: | Kumeta, H, Kobashigawa, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2008-12-07 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method
J.Biomol.Nmr, 43, 2009
|
|
2KNB
 
 | | Solution NMR structure of the parkin Ubl domain in complex with the endophilin-A1 SH3 domain | | Descriptor: | E3 ubiquitin-protein ligase parkin, Endophilin-A1 | | Authors: | Trempe, J, Guennadi, K, Edna, C.M, Kalle, G. | | Deposit date: | 2009-08-20 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination.
Mol.Cell, 36, 2009
|
|
2L3S
 
 | | Structure of the autoinhibited Crk | | Descriptor: | Autoinhibited Crk protein | | Authors: | Kalodimos, C.G, Sarkar, P, Saleh, T, Tzeng, S.R, Birge, R. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for regulation of the Crk signaling protein by a proline switch.
Nat.Chem.Biol., 7, 2011
|
|
