2AAY
 
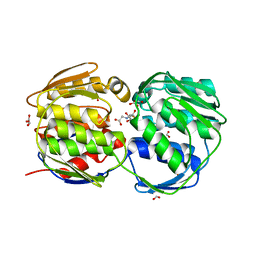 | | EPSP synthase liganded with shikimate and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, ... | | Authors: | Priestman, M.A, Healy, M.L, Funke, T, Becker, A. | | Deposit date: | 2005-07-14 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis for the glyphosate-insensitivity of the reaction of 5-enolpyruvylshikimate 3-phosphate synthase with shikimate.
Febs Lett., 579, 2005
|
|
1UAE
 
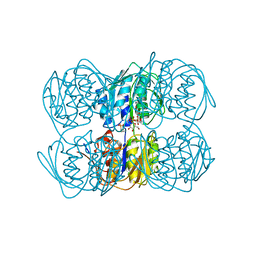 | | STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, [(1R)-1-hydroxypropyl]phosphonic acid | | Authors: | Skarzynski, T. | | Deposit date: | 1996-09-30 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Structure, 4, 1996
|
|
6Q03
 
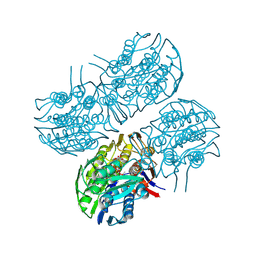 | | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine) | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine)
To Be Published
|
|
6Q0A
 
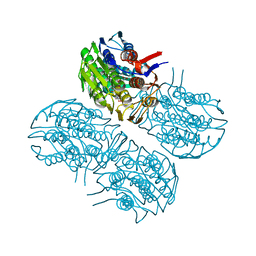 | | Crystal structure of MurA from Clostridium difficile, mutation C116D, n the presence of UDP-N-acetylmuramic acid | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutation C116D, n the presence of UDP-N-acetylmuramic acid
To Be Published
|
|
6Q11
 
 | | Crystal structure of MurA from Clostridium difficile, mutation C116S, in the presence of URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutation C116S, in the presence of URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID
To Be Published
|
|
6Q0Y
 
 | | Crystal structure of MurA from Clostridium difficile, mutant C116S, in the presence of Uridine-Diphosphate-N-Acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutant C116S, in the presence of Uridine-Diphosphate-N-Acetylglucosamine
To Be Published
|
|
2BJB
 
 | | Mycobacterium Tuberculosis Epsp Synthase In Unliganded State | | Descriptor: | 3-PHOSPHOSHIKIMATE 1-CARBOXYVINYLTRANSFERASE, ACETATE ION, SODIUM ION | | Authors: | Bourenkov, G.P, Kachalova, G.S, Strizhov, N, Bruning, M, Vagin, A, Bartunik, H.D. | | Deposit date: | 2005-02-01 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium Tuberculosis Epsp Synthase in Unliganded State
To be Published
|
|
1X8T
 
 | | EPSPS liganded with the (R)-phosphonate analog of the tetrahedral reaction intermediate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, [3R-[3A,4A,5B(R*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Priestman, M.A, Healy, M.L, Becker, A, Alberg, D.G, Bartlett, P.A, Schonbrunn, E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction of phosphonate analogues of the tetrahedral reaction intermediate with 5-enolpyruvylshikimate-3-phosphate synthase in atomic detail.
Biochemistry, 44, 2005
|
|
1X8R
 
 | | EPSPS liganded with the (S)-phosphonate analog of the tetrahedral reaction intermediate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, [3R-[3A,4A,5B(S*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Priestman, M.A, Healy, M.L, Becker, A, Alberg, D.G, Bartlett, P.A, Schonbrunn, E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of phosphonate analogues of the tetrahedral reaction intermediate with 5-enolpyruvylshikimate-3-phosphate synthase in atomic detail.
Biochemistry, 44, 2005
|
|
3FK0
 
 | | E. coli EPSP synthase (TIPS mutation) liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3FJX
 
 | | E. coli EPSP synthase (T97I) liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3FK1
 
 | | E. coli EPSP synthase (TIPS mutation) liganded with S3P and glyphosate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, N-(phosphonomethyl)glycine, ... | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3FJZ
 
 | | E. coli EPSP synthase (T97I) liganded with S3P and glyphosate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, N-(phosphonomethyl)glycine, ... | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
1YBG
 
 | | MurA inhibited by a derivative of 5-sulfonoxy-anthranilic acid | | Descriptor: | N-METHYL-N-{2-[(2-NAPHTHYLSULFONYL)AMINO]-5-[(2-NAPHTHYLSULFONYL)OXY]BENZOYL}-L-ASPARTIC ACID, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Priestman, M.A, Abdul-Latif, F.A, Delachaume, C, Fassy, F, Schonbrunn, E. | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Inhibitor That Suspends the Induced Fit Mechanism of UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA).
J.Biol.Chem., 280, 2005
|
|
2GGA
 
 | | CP4 EPSP synthase liganded with S3P and Glyphosate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, GLYPHOSATE, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E, Funke, T. | | Deposit date: | 2006-03-23 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis for the herbicide resistance of Roundup Ready crops.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3ISS
 
 | | Crystal structure of enolpyruvyl-UDP-GlcNAc synthase (MurA):UDP-N-acetylmuramic acid:phosphite from Escherichia coli | | Descriptor: | PHOSPHITE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Jackson, S.G, Zhang, F, Chindemi, P, Junop, M.S, Berti, P.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence of Kinetic Control of Ligand Binding and Staged Product Release in MurA (Enolpyruvyl UDP-GlcNAc Synthase)-Catalyzed Reactions .
Biochemistry, 48, 2009
|
|
3KQA
 
 | | MurA dead-end complex with terreic acid | | Descriptor: | (5S)-2,5-dihydroxy-3-methylcyclohex-2-ene-1,4-dione, CALCIUM ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Schonbrunn, E. | | Deposit date: | 2009-11-17 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) .
Biochemistry, 49, 2010
|
|
3KQJ
 
 | | MurA binary complex with UDP-N-acetylglucosamine | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Schonbrunn, E. | | Deposit date: | 2009-11-17 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Natural Product Antibiotic Terreic Acid is a Mechanism-Based Inhibitor of the Bacterial Enzyme MurA in vitro but not in vivo.
To be Published
|
|
3KR6
 
 | | MurA dead-end complex with fosfomycin | | Descriptor: | UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, [(1R)-1-hydroxypropyl]phosphonic acid | | Authors: | Schonbrunn, E. | | Deposit date: | 2009-11-17 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) .
Biochemistry, 49, 2010
|
|
3LTH
 
 | | E. cloacae MurA dead-end complex with UNAG and fosfomycin | | Descriptor: | UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, [(1R)-1-hydroxypropyl]phosphonic acid | | Authors: | Schonbrunn, E. | | Deposit date: | 2010-02-15 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) .
Biochemistry, 49, 2010
|
|
7TM6
 
 | |
7TM4
 
 | |
7TM5
 
 | |
1DLG
 
 | | CRYSTAL STRUCTURE OF THE C115S ENTEROBACTER CLOACAE MURA IN THE UN-LIGANDED STATE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE MURA | | Authors: | Schonbrunn, E, Eschenburg, S, Krekel, F, Luger, K, Amrhein, N. | | Deposit date: | 1999-12-09 | | Release date: | 2000-04-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the loop containing residue 115 in the induced-fit mechanism of the bacterial cell wall biosynthetic enzyme MurA.
Biochemistry, 39, 2000
|
|
1EJD
 
 | | Crystal structure of unliganded mura (type1) | | Descriptor: | CYCLOHEXYLAMMONIUM ION, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
