3D9X
 
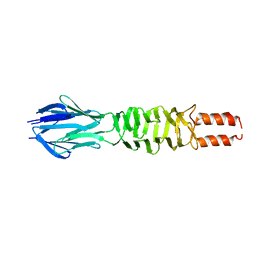 | |
2FGR
 
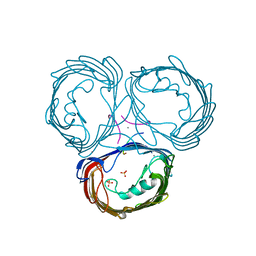 | | High resolution Xray structure of Omp32 | | Descriptor: | CALCIUM ION, Outer membrane porin protein 32, PAP, ... | | Authors: | Zeth, K, Zachariae, U, Engelhardt, H. | | Deposit date: | 2005-12-22 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution crystal structures and molecular dynamics studies reveal substrate binding in the porin omp32
J.Biol.Chem., 281, 2006
|
|
1TK6
 
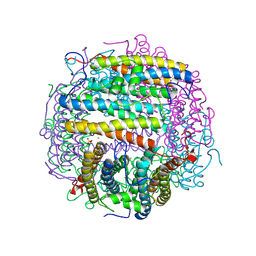 | | Iron-oxo clusters biomineralizing on protein surfaces. Structural analysis of H.salinarum DpsA in its low and high iron states | | Descriptor: | FE (III) ION, Iron-rich dpsA-homolog protein, MAGNESIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2004-06-08 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TKP
 
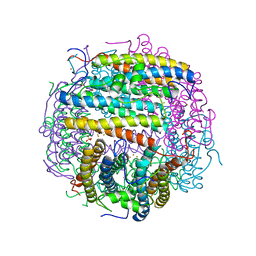 | | Iron-oxo clusters biomineralizing on protein surfaces. Structural analysis of H.salinarum DpsA in its low and high iron states | | Descriptor: | FE (III) ION, Iron-rich dpsA-homolog protein, SODIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TJO
 
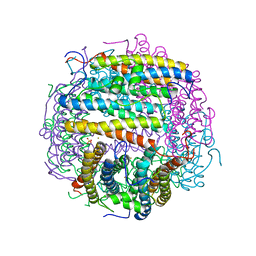 | | Iron-oxo clusters biomineralizing on protein surfaces. Structural analysis of H.salinarum DpsA in its low and high iron states | | Descriptor: | FE (III) ION, Iron-rich dpsA-homolog protein, MAGNESIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2004-06-07 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TKO
 
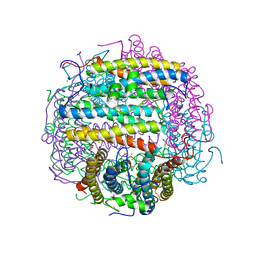 | | Iron-oxo clusters biomineralizing on protein surfaces. Structural analysis of H.salinarum DpsA in its low and high iron states | | Descriptor: | FE (III) ION, Iron-rich dpsA-homolog protein, SODIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3DA4
 
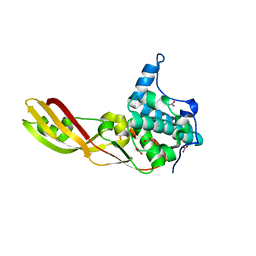 | | Crystal Structure of Colicin M, a Novel Phosphatase Specifically Imported by Escherichia Coli | | Descriptor: | Colicin-M, NITRATE ION | | Authors: | Zeth, K, Albrecht, R, Romer, C, Braun, V. | | Deposit date: | 2008-05-28 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli
J.Biol.Chem., 283, 2008
|
|
3DA3
 
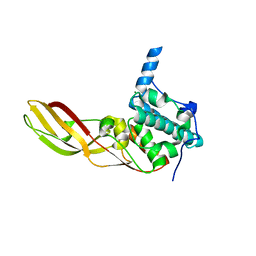 | | Crystal Structure of Colicin M, A Novel Phosphatase Specifically Imported by Escherichia Coli | | Descriptor: | Colicin-M, MAGNESIUM ION | | Authors: | Zeth, K, Albrecht, R, Romer, C, Braun, V. | | Deposit date: | 2008-05-28 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli
J.Biol.Chem., 283, 2008
|
|
3ZX6
 
 | | Structure of Hamp(AF1503)-Tsr fusion - Hamp (A291V) mutant | | Descriptor: | CHLORIDE ION, HAMP, METHYL-ACCEPTING CHEMOTAXIS PROTEIN I | | Authors: | Zeth, K, Ferris, H.U, Lupas, A.L. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Axial Helix Rotation as a Mechanism for Signal Regulation Inferred from the Crystallographic Analysis of the E. Coli Serine Chemoreceptor.
J.Struct.Biol., 186, 2014
|
|
3ZCC
 
 | |
3ZRX
 
 | |
3ZRW
 
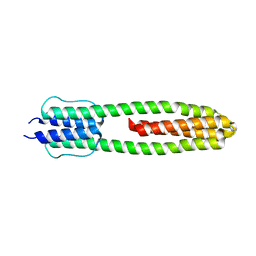 | | The structure of the dimeric Hamp-Dhp fusion A291V mutant | | Descriptor: | AF1503 PROTEIN, OSMOLARITY SENSOR PROTEIN ENVZ | | Authors: | Zeth, K, Hulko, M, Ferris, H.U, Martin, J, Lupas, A.N. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
3ZRV
 
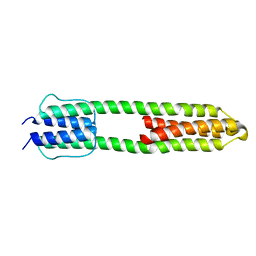 | | The high resolution structure of a dimeric Hamp-Dhp fusion displays asymmetry - A291F mutant | | Descriptor: | HAMP, OSMOLARITY SENSOR PROTEIN ENVZ | | Authors: | Zeth, K, Hulko, M, Ferris, H.U, Martin, J. | | Deposit date: | 2011-06-20 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
4C3K
 
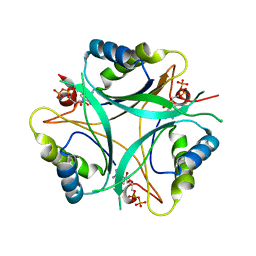 | | Structure of mixed PII-ADP complexes from S. elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NITROGEN REGULATORY PROTEIN P-II, PHOSPHATE ION | | Authors: | Zeth, K, Forchhammer, K. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural Basis and Target-Specific Modulation of Adp Sensing by the Synechococcus Elongatus Pii Signaling Protein.
J.Biol.Chem., 289, 2014
|
|
4C3M
 
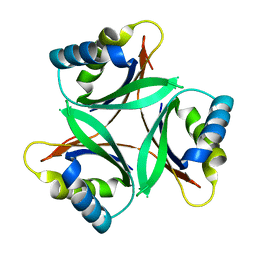 | |
4C4V
 
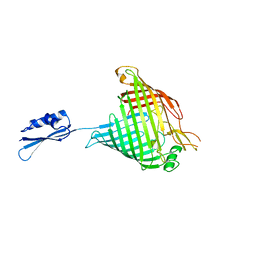 | |
4C3L
 
 | | Structure of wildtype PII from S. elongatus at high resolution | | Descriptor: | CHLORIDE ION, NITROGEN REGULATORY PROTEIN P-II, TETRAETHYLENE GLYCOL | | Authors: | Zeth, K, Forchhammer, K. | | Deposit date: | 2013-08-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis and Target-Specific Modulation of Adp Sensing by the Synechococcus Elongatus Pii Signaling Protein.
J.Biol.Chem., 289, 2014
|
|
4AFF
 
 | | High resolution structure of a PII mutant (I86N) protein in complex with ATP, MG and FLC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRATE ANION, MAGNESIUM ION, ... | | Authors: | Zeth, K, Fokina, O, Chellamuthu, V.R, Forchhammer, K. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | An Engineered Pii Protein Variant that Senses a Novel Ligand: Atomic Resolution Structure of the Complex with Citrate.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4CSO
 
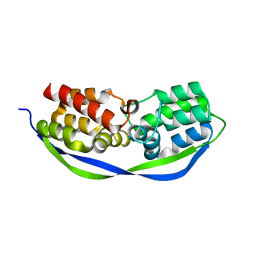 | | The structure of OrfY from Thermoproteus tenax | | Descriptor: | ORFY PROTEIN, TRANSCRIPTION FACTOR | | Authors: | Zeth, K, Hagemann, A, Siebers, B, Martin, J, Lupas, A.N. | | Deposit date: | 2014-03-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
4AUI
 
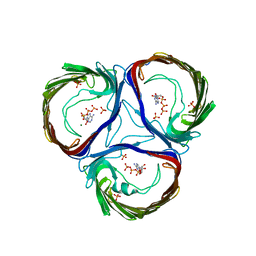 | | STRUCTURE AND FUNCTION OF THE PORB PORIN FROM DISSEMINATING N. GONORRHOEAE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Zeth, K, Kozjak-Pavlovic, V, Faulstich, M, Hurwitz, R, Kepp, O, Rudel, T. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Function of the Porb Porin from Disseminating Neisseria Gonorrhoeae.
Biochem.J., 449, 2013
|
|
4BAX
 
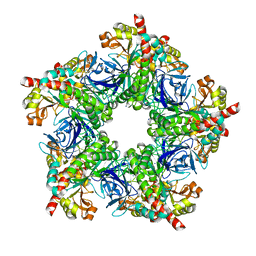 | |
4AEQ
 
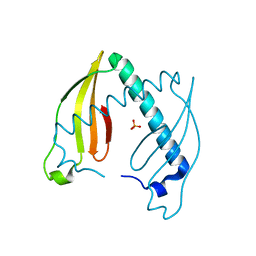 | | Crystal structure of the dimeric immunity protein Cmi solved by direct methods (Arcimboldo) | | Descriptor: | COLICIN-M IMMUNITY PROTEIN, PHOSPHATE ION | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Uson, I, Braun, V. | | Deposit date: | 2012-01-12 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | The Crystal Structure of the Dimeric Colicin M Immunity
J.Struct.Biol., 178, 2012
|
|
4ARJ
 
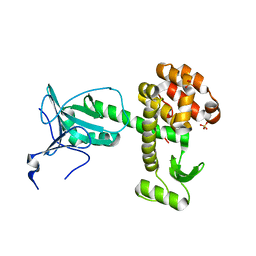 | | Crystal structure of a pesticin (translocation and receptor binding domain) from Y. pestis and T4-lysozyme chimera | | Descriptor: | PESTICIN, LYSOZYME, SULFATE ION | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARM
 
 | | Structure of the inactive pesticin T201A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARQ
 
 | | Structure of the pesticin S89C, S285C double mutant | | Descriptor: | MAGNESIUM ION, PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
