1J0G
 
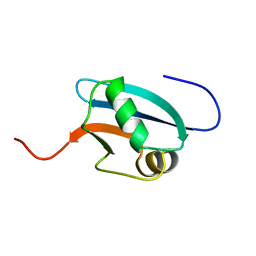 | | Solution Structure of Mouse Hypothetical 9.1 kDa Protein, A Ubiquitin-like Fold | | Descriptor: | Hypothetical Protein 1810045K17 | | Authors: | Zhao, C, Kigawa, T, Koshiba, S, Tochio, N, Kobayashi, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical 9.1 kDa Protein, A Ubiquitin-like Fold
To be Published
|
|
1J1U
 
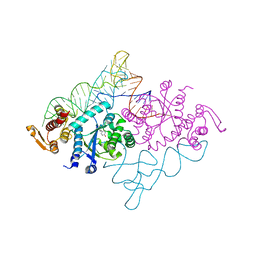 | | Crystal structure of archaeal tyrosyl-tRNA synthetase complexed with tRNA(Tyr) and L-tyrosine | | Descriptor: | MAGNESIUM ION, TYROSINE, Tyrosyl-tRNA synthetase, ... | | Authors: | Kobayashi, T, Nureki, O, Ishitani, R, Tukalo, M, Cusack, S, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-17 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion
NAT.STRUCT.BIOL., 10, 2003
|
|
1J27
 
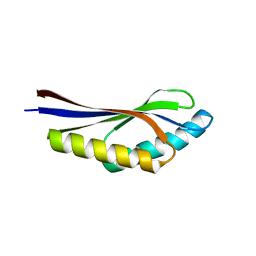 | | Crystal structure of a hypothetical protein, TT1725, from Thermus thermophilus HB8 at 1.7A resolution | | Descriptor: | hypothetical protein TT1725 | | Authors: | Seto, A, Shirouzu, M, Terada, T, Murayama, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-26 | | Release date: | 2003-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a hypothetical protein, TT1725, from Thermus thermophilus HB8 at 1.7 A resolution
Proteins, 53, 2003
|
|
1J2W
 
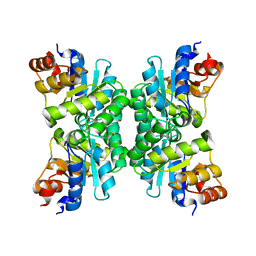 | | Tetrameric Structure of aldolase from Thermus thermophilus HB8 | | Descriptor: | Aldolase protein | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2HOQ
 
 | |
2HOW
 
 | |
2IC7
 
 | | Crystal Structure of Maltose Transacetylase from Geobacillus kaustophilus | | Descriptor: | Maltose transacetylase | | Authors: | Liu, Z.J, Li, Y, Chen, L, Zhu, J, Rose, J.P, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-12 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Maltose Transacetylase From Geobacillus kaustophilus at 1.78 Angstrom Resolution
To be Published
|
|
2ICU
 
 | | Crystal Structure of Hypothetical Protein YedK From Escherichia coli | | Descriptor: | Hypothetical protein yedK | | Authors: | Chen, L, Liu, Z.J, Li, Y, Zhao, M, Rose, J, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-13 | | Release date: | 2006-11-07 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Hypothetical Protein YedK From Escherichia coli
To be Published
|
|
2J40
 
 | | 1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound inhibitor L-proline and NAD. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Sakamoto, K, Nishio, M, Yokoyama, S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Ternary Complex of Delta1-Pyrroline-5-Carboxylate Dehydrogenase with Substrate Mimic and Co-Factoer
To be Published
|
|
2IY6
 
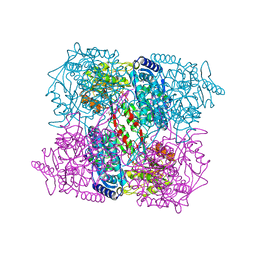 | | 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE FROM THERMUS WITH BOUND CITRATE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Sakamoto, K, Nishio, M, Yokoyama, S. | | Deposit date: | 2006-07-13 | | Release date: | 2006-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
2J5N
 
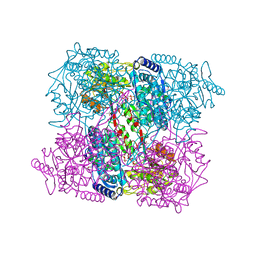 | | 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE FROM THERMUS THERMOPHIRUS WITH BOUND INHIBITOR GLYCINE AND NAD. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Sakamoto, K, Nishio, M, Yokoyama, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of Ternary Complex of Delta1-Pyrroline-5-Carboxylate Dehydrogenase with Substrate Mimic and Co-Factoer
To be Published
|
|
2JNS
 
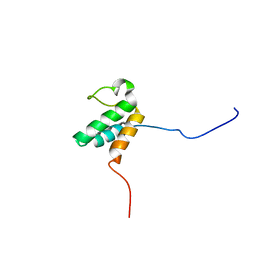 | |
2KBO
 
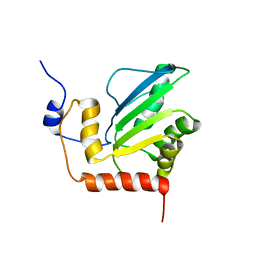 | | Structure, interaction, and real-time monitoring of the enzymatic reaction of wild type APOBEC3G | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Furukawa, A, Nagata, T, Matsugami, A, Habu, Y, Sugiyama, R, Hayashi, F, Kobayashi, N, Yokoyama, S, Takaku, H, Katahira, M. | | Deposit date: | 2008-12-04 | | Release date: | 2009-02-03 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G
Embo J., 28, 2009
|
|
1IYF
 
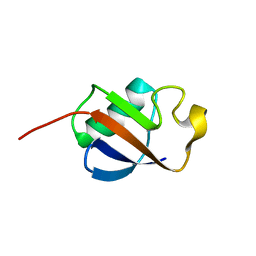 | | Solution structure of ubiquitin-like domain of human parkin | | Descriptor: | parkin | | Authors: | Sakata, E, Yamaguchi, Y, Kurimoto, E, Kikuchi, J, Yokoyama, S, Kawahara, H, Yokosawa, H, Hattori, N, Mizuno, Y, Tanaka, K, Kato, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain
EMBO REP., 4, 2003
|
|
1IVZ
 
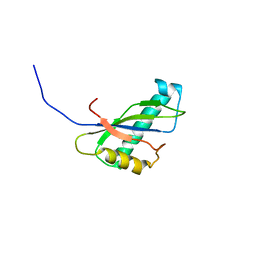 | | Solution structure of the SEA domain from murine hypothetical protein homologous to human mucin 16 | | Descriptor: | hypothetical protein 1110008I14RIK | | Authors: | Maeda, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-02 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16)
J.Biol.Chem., 279, 2004
|
|
3VU8
 
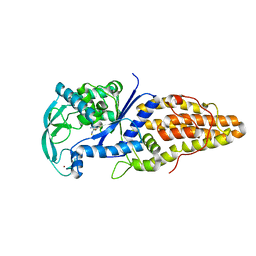 | | Metionyl-tRNA synthetase from Thermus thermophilus complexed with methionyl-adenylate analogue | | Descriptor: | Methionine--tRNA ligase, N-[METHIONYL]-N'-[ADENOSYL]-DIAMINOSULFONE, ZINC ION | | Authors: | Konno, M, Kato-Murayama, M, Toma-Fukai, S, Uchikawa, E, Nureki, O, Yokoyama, S. | | Deposit date: | 2012-06-22 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The modeling of structures of specific conformation of homosysteine-AMP leading to thiolactone-formation on class Ia aminoacyl-tRNA synthetases
To be Published
|
|
1IV0
 
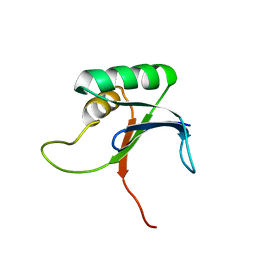 | |
5X60
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 9 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
2OM6
 
 | | Hypothetical Protein (Probable Phosphoserine Phosph (PH0253) from Pyrococcus Horikoshii OT3 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable phosphoserine phosphatase, ... | | Authors: | Jeyakanthan, J, Vaijayanthimala, S, Gayathri, D, Velmurugan, D, Baba, S, Ebihara, A, Shinkai, A, Kuramitsu, S, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-21 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hypothetical Protein (Probable Phosphoserine Phosph (PH0253) from Pyrococcus Horikoshii OT3
To be Published
|
|
4YZO
 
 | | Crystal Structure Analysis of Thiolase-like protein, ST0096 from Sulfolobus Tokodaii | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Padmanabhan, B, Manjula, R, Yokoyama, S, Bessho, Y. | | Deposit date: | 2015-03-25 | | Release date: | 2016-03-30 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Analysis of Thiolase-like protein, ST0096 from Sulfolobus Tokodaii
To Be Published
|
|
2PBQ
 
 | | Crystal structure of molybdenum cofactor biosynthesis (aq_061) From aquifex aeolicus VF5 | | Descriptor: | Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Mahesh, S, Kanaujia, S.P, Ramakumar, S, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of molybdenum cofactor biosynthesis (aq_061) from aquifex aeolicus VF5
to be published
|
|
3VR2
 
 | | Crystal structure of nucleotide-free A3B3 complex from Enterococcus hirae V-ATPase [eA3B3] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR4
 
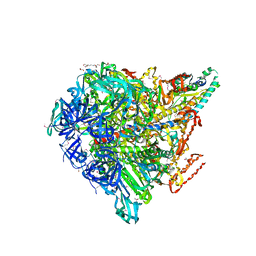 | | Crystal structure of Enterococcus hirae V1-ATPase [eV1] | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR6
 
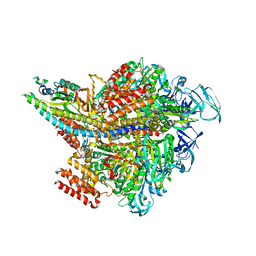 | | Crystal structure of AMP-PNP bound Enterococcus hirae V1-ATPase [bV1] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR3
 
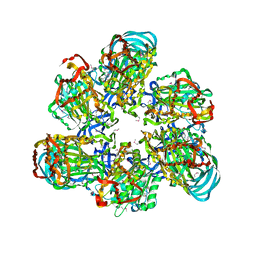 | | Crystal structure of AMP-PNP bound A3B3 complex from Enterococcus hirae V-ATPase [bA3B3] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
