3A1Q
 
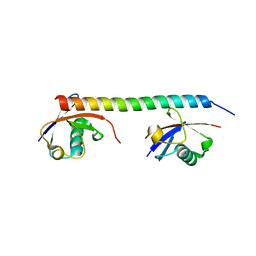 | | Crystal structure of the mouse RAP80 UIMs in complex with Lys63-linked di-ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin interaction motif-containing protein 1 | | Authors: | Sato, Y, Yoshikawa, A, Mimura, H, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-04-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80
Embo J., 28, 2009
|
|
3A9K
 
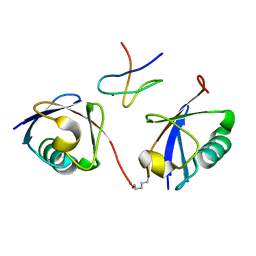 | | Crystal structure of the mouse TAB3-NZF in complex with Lys63-linked di-ubiquitin | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3, Ubiquitin, ZINC ION | | Authors: | Sato, Y, Yoshikawa, A, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3
Embo J., 28, 2009
|
|
3B0A
 
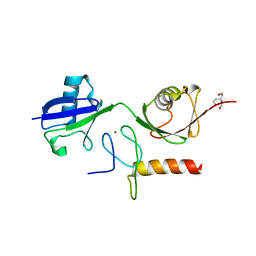 | | Crystal structure of the mouse HOIL1-L-NZF in complex with linear di-ubiquitin | | Descriptor: | Polyubiquitin-C, RanBP-type and C3HC4-type zinc finger-containing protein 1, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | Sato, Y, Fujita, H, Yoshikawa, A, Yamashita, M, Yamagata, A, Kaiser, S.E, Iwai, K, Fukai, S. | | Deposit date: | 2011-06-07 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B2E
 
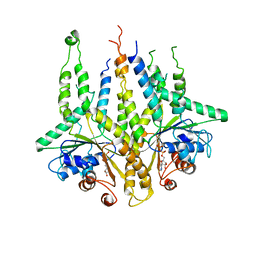 | |
6J6X
 
 | | Crystal structure of apo GGTaseIII | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Geranylgeranyl transferase type-2 subunit beta, MAGNESIUM ION, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2019-01-16 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.962 Å) | | Cite: | Crystal structure of apo GGTaseIII
To Be Published
|
|
6J74
 
 | | Complex of GGTaseIII and full-length Ykt6 | | Descriptor: | Geranylgeranyl transferase type-2 subunit beta, PHOSPHATE ION, Protein prenyltransferase alpha subunit repeat-containing protein 1, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2019-01-16 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.212 Å) | | Cite: | Complex of GGTaseIII and full-length Ykt6
To Be Published
|
|
6J7F
 
 | | Complex of GGTaseIII, farnesyl-Ykt6 (C-terminal methylated), and GGPP | | Descriptor: | DIPHOSPHATE, FARNESYL, GERAN-8-YL GERAN, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Complex of GGTaseIII, farnesyl-Ykt6 (C-terminal methylated), and GGPP
To Be Published
|
|
6J7X
 
 | | Complex of GGTaseIII, farnesyl-Ykt6, and GGPP | | Descriptor: | FORMIC ACID, GERANYLGERANYL DIPHOSPHATE, Geranylgeranyl transferase type-2 subunit beta, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Complex of GGTaseIII, farnesyl-Ykt6 (C-terminal methylated), and GGPP
To Be Published
|
|
