1D6K
 
 | | NMR SOLUTION STRUCTURE OF THE 5S RRNA E-LOOP/L25 COMPLEX | | Descriptor: | 5S RRNA E-LOOP (5SE), RIBOSOMAL PROTEIN L25 | | Authors: | Stoldt, M, Wohnert, J, Ohlenschlager, O, Gorlach, M, Brown, L.R. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-22 | | Last modified: | 2022-03-23 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the 5S rRNA E-domain-protein L25 complex shows preformed and induced recognition.
EMBO J., 18, 1999
|
|
1KMA
 
 | | NMR Structure of the Domain-I of the Kazal-type Thrombin Inhibitor Dipetalin | | Descriptor: | DIPETALIN | | Authors: | Schlott, B, Wohnert, J, Icke, C, Hartmann, M, Ramachandran, R, Guhrs, K.-H, Glusa, E, Flemming, J, Gorlach, M, Grosse, F, Ohlenschlager, O. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Interaction of Kazal-type inhibitor domains with serine proteinases: biochemical and structural studies.
J.Mol.Biol., 318, 2002
|
|
1RFR
 
 | | NMR structure of the 30mer stemloop-D of coxsackieviral RNA | | Descriptor: | stemloop-D RNA of the 5'-cloverleaf of coxsackievirus B3 | | Authors: | Ohlenschlager, O, Wohnert, J, Bucci, E, Seitz, S, Hafner, S, Ramachandran, R, Zell, R, Gorlach, M. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-23 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf
RNA and its interaction with the proteinase 3C.
STRUCTURE, 12, 2004
|
|
3BBD
 
 | | M. jannaschii Nep1 complexed with S-adenosyl-homocysteine | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
3BBH
 
 | | M. jannaschii Nep1 complexed with Sinefungin | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, SINEFUNGIN | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
3BBE
 
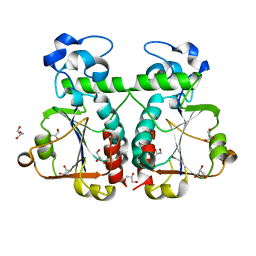 | | M. jannaschii Nep1 | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
2KXM
 
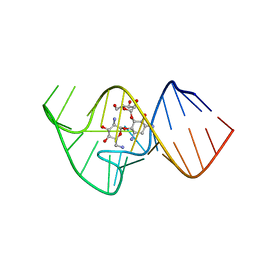 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostmycin complex | | Descriptor: | RIBOSTAMYCIN, RNA (27-MER) | | Authors: | Duchardt-Ferner, E, Weigand, J.E, Ohlenschlager, O, Schmidtke, S.R, Suess, B, Wohnert, J. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2014-02-05 | | Method: | SOLUTION NMR | | Cite: | Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6GZR
 
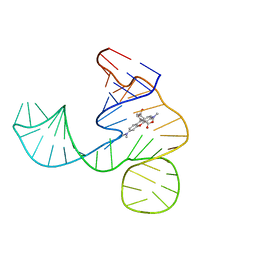 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, tetramethylrhodamine aptamer | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-17 | | Last modified: | 2020-01-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
6GZK
 
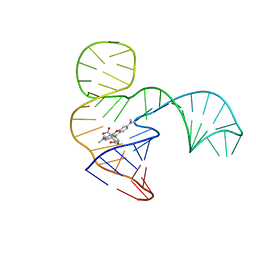 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, TMR3 (48-MER) | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-04 | | Release date: | 2019-07-17 | | Last modified: | 2020-01-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
2MXS
 
 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | Descriptor: | PAROMOMYCIN, RNA (27-MER) | | Authors: | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | Deposit date: | 2015-01-14 | | Release date: | 2015-12-09 | | Last modified: | 2016-02-03 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
