4BIA
 
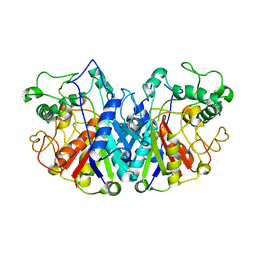 | | Crystal structure of SCP2 thiolase from Trypanosoma brucei: The C337A mutant. | | Descriptor: | 3-KETOACYL-COA THIOLASE, PUTATIVE | | Authors: | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
4BI9
 
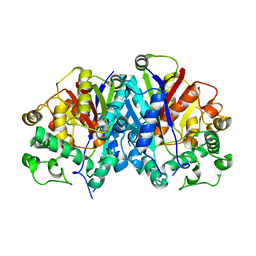 | | Crystal structure of wild-type SCP2 thiolase from Trypanosoma brucei. | | Descriptor: | 3-KETOACYL-COA THIOLASE, PUTATIVE | | Authors: | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
1QDS
 
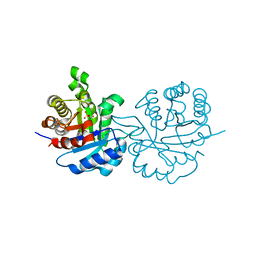 | | SUPERSTABLE E65Q MUTANT OF LEISHMANIA MEXICANA TRIOSEPHOSPHATE ISOMERASE (TIM) | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Lambeir, A.M, Backmann, J, Ruiz-Sanz, J, Filimonov, V, Nielsen, J.E, Vriend, G, Kursula, I, Norledge, B.V, Wierenga, R.K. | | Deposit date: | 1999-07-10 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The ionization of a buried glutamic acid is thermodynamically linked to the stability of Leishmania mexicana triose phosphate isomerase.
Eur.J.Biochem., 267, 2000
|
|
1SHG
 
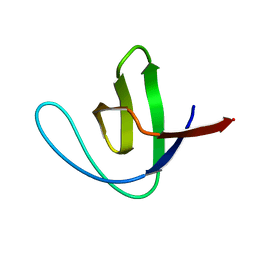 | | CRYSTAL STRUCTURE OF A SRC-HOMOLOGY 3 (SH3) DOMAIN | | Descriptor: | ALPHA-SPECTRIN SH3 DOMAIN | | Authors: | Noble, M, Pauptit, R, Musacchio, A, Saraste, M, Wierenga, R.K. | | Deposit date: | 1993-05-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Src-homology 3 (SH3) domain.
Nature, 359, 1992
|
|
1GYR
 
 | | Mutant form of enoyl thioester reductase from Candida tropicalis | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, GLYCEROL, SULFATE ION | | Authors: | Airenne, T.T, Torkko, J.M, Van Der Plas, S, Sormunen, R.T, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-04-29 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1GUF
 
 | | Enoyl thioester reductase from Candida tropicalis | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1, MITOCHONDRIAL, ... | | Authors: | Airenne, T.T, Torkko, J.M, Van Der Plas, S, Sormunen, R.T, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-01-25 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1NL7
 
 | |
1TJC
 
 | | Crystal structure of peptide-substrate-binding domain of human type I collagen prolyl 4-hydroxylase | | Descriptor: | Prolyl 4-hydroxylase alpha-1 subunit | | Authors: | Pekkala, M, Hieta, R, Bergmann, U, Kivirikko, K.I, Wierenga, R.K, Myllyharju, J. | | Deposit date: | 2004-06-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Peptide-Substrate-binding Domain of Collagen Prolyl 4-Hydroxylases Is a Tetratricopeptide Repeat Domain with Functional Aromatic Residues.
J.Biol.Chem., 279, 2004
|
|
1DLU
 
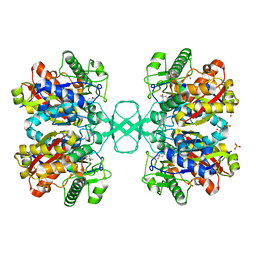 | | UNLIGANDED BIOSYNTHETIC THIOLASE FROM ZOOGLOEA RAMIGERA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BIOSYNTHETIC THIOLASE, SULFATE ION | | Authors: | Modis, Y, Wierenga, R.K. | | Deposit date: | 1999-12-12 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase.
J.Mol.Biol., 297, 2000
|
|
1DM3
 
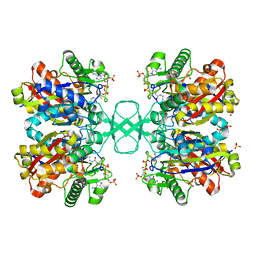 | |
1DLV
 
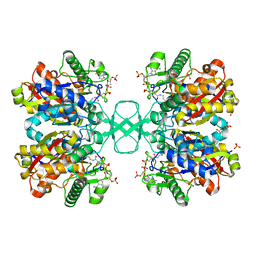 | |
1OU6
 
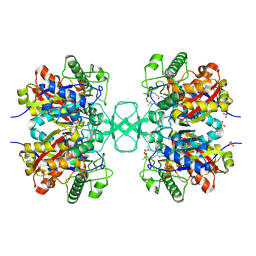 | |
1GU7
 
 | | Enoyl thioester reductase from Candida tropicalis | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1,MITOCHONDRIAL, GLYCEROL, ... | | Authors: | Airenne, T.T, Torkko, J.M, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1HNU
 
 | | CRYSTAL STRUCTURE OF PEROXISOMAL DELTA3-DELTA2-ENOYL-COA ISOMERASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 1,2-ETHANEDIOL, D3,D2-ENOYL COA ISOMERASE ECI1, PERRHENATE | | Authors: | Mursula, A.M, van Aalten, D.M.F, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2000-12-08 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of delta(3)-delta(2)-enoyl-CoA isomerase.
J.Mol.Biol., 309, 2001
|
|
1HNO
 
 | | CRYSTAL STRUCTURE OF PEROXISOMAL DELTA3-DELTA2-ENOYL-COA ISOMERASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 1,2-ETHANEDIOL, D3,D2-ENOYL COA ISOMERASE ECI1 | | Authors: | Mursula, A.M, van Aalten, D.M.F, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2000-12-08 | | Release date: | 2001-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of delta(3)-delta(2)-enoyl-CoA isomerase.
J.Mol.Biol., 309, 2001
|
|
1KV5
 
 | | Structure of Trypanosoma brucei brucei TIM with the salt-bridge-forming residue Arg191 mutated to Ser | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, ... | | Authors: | Kursula, I, Partanen, S, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-01-25 | | Release date: | 2002-03-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The importance of the conserved Arg191-Asp227 salt bridge of triosephosphate isomerase for folding, stability, and catalysis
FEBS Lett., 518, 2002
|
|
1SQ7
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SPQ
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SSG
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SSD
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SW0
 
 | | Triosephosphate isomerase from Gallus gallus, loop 6 hinge mutant K174L, T175W | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SW3
 
 | | Triosephosphate isomerase from Gallus gallus, loop 6 mutant T175V | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SU5
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SW7
 
 | | Triosephosphate isomerase from Gallus gallus, loop 6 mutant K174N, T175S, A176S | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1IIG
 
 | | STRUCTURE OF TRYPANOSOMA BRUCEI BRUCEI TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH 3-PHOSPHONOPROPIONATE | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E, Wierenga, R.K, Lambeir, A.M, Opperdoes, F.R, Thunnissen, A.M, Kalk, K.H, Groendijk, H, Hol, W.G.J. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes.
Proteins, 10, 1991
|
|
