4DUU
 
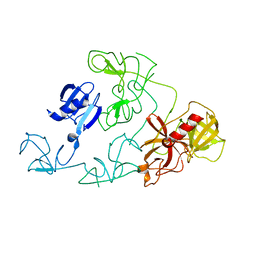 | |
4OV8
 
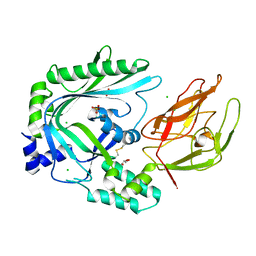 | | Crystal Structure of the TMH1-lock mutant of the mature form of pleurotolysin B | | Descriptor: | CHLORIDE ION, GLYCEROL, Pleurotolysin B, ... | | Authors: | Kondos, S.C, Law, R.H.P, Whisstock, J.C, Dunstone, M.A. | | Deposit date: | 2014-02-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin
Plos Biol., 13, 2015
|
|
4OEB
 
 | | Structure of membrane binding protein pleurotolysin A from Pleurotus ostreatus | | Descriptor: | Pleurotolysin A, SULFATE ION | | Authors: | Dunstone, M.A, Caradoc-Davies, T.T, Whisstock, J.C, Law, R.H.P. | | Deposit date: | 2014-01-12 | | Release date: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
3NSJ
 
 | | The X-ray crystal structure of lymphocyte perforin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Law, R.H, Whisstock, J.C, Caradoc-Davies, T.T. | | Deposit date: | 2010-07-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structural basis for membrane binding and pore formation by lymphocyte perforin.
Nature, 468, 2010
|
|
3VP6
 
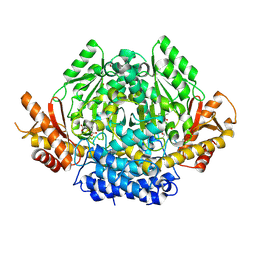 | | Structural characterization of Glutamic Acid Decarboxylase; insights into the mechanism of autoinactivation | | Descriptor: | 4-oxo-4H-pyran-2,6-dicarboxylic acid, GLYCEROL, Glutamate decarboxylase 1 | | Authors: | Langendorf, C.G, Tuck, K.L, Key, T.L.G, Rosado, C.J, Wong, A.S.M, Fenalti, G, Buckle, A.M, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the mechanism through which human glutamic acid decarboxylase auto-activates
Biosci.Rep., 33, 2013
|
|
4F88
 
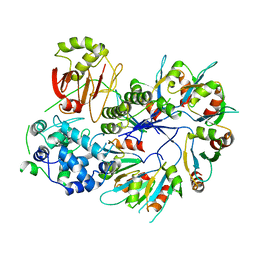 | | X-ray Crystal Structure of PlyC | | Descriptor: | PlyCA, PlyCB | | Authors: | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F87
 
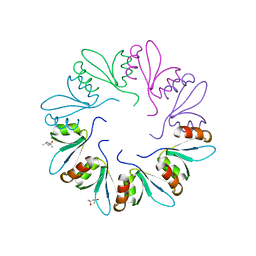 | | X-ray Crystal Structure of PlyCB | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3JZ4
 
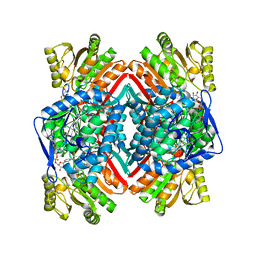 | | Crystal structure of E. coli NADP dependent enzyme | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase [NADP+] | | Authors: | Langendorf, C.G, Key, T.L.G, Fenalti, G, Kan, W.T, Buckle, A.M, Caradoc-Davies, T, Tuck, K.L, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2009-09-22 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structure of Escherichia coli succinic semialdehyde dehydrogenase; structural insights into NADP+/enzyme interactions.
Plos One, 5, 2010
|
|
6O1Z
 
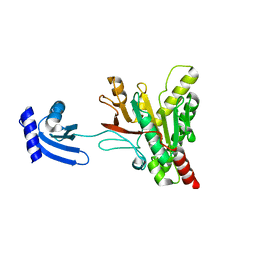 | | Structure of pCW3 conjugation coupling protein TcpA hexagonal crystal form | | Descriptor: | DNA translocase coupling protein | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
6O1Y
 
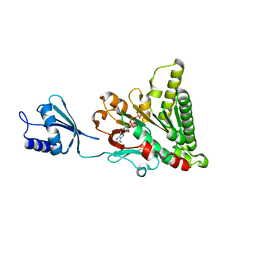 | | Structure of pCW3 conjugation coupling protein TcpA monomeric form with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA translocase coupling protein, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
6O1W
 
 | | Structure of pCW3 conjugation coupling protein TcpA monomer orthorhombic crystal form | | Descriptor: | DNA translocase coupling protein, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
6O1X
 
 | | Structure of pCW3 conjugation coupling protein TcpA monomer form with ATPgS | | Descriptor: | DNA translocase coupling protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
4P0O
 
 | | Cleaved Serpin 42Da | | Descriptor: | Serine protease inhibitor 4, isoform B | | Authors: | Ellisdon, A.M, Whisstock, J.C. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of cleaved Serpin 42 Da from Drosophila melanogaster.
Bmc Struct.Biol., 14, 2014
|
|
4P0F
 
 | | Cleaved Serpin 42Da (C 2 2 21) | | Descriptor: | Serine protease inhibitor 4, isoform B | | Authors: | Ellisdon, A.M, Whisstock, J.C. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution structure of cleaved Serpin 42 Da from Drosophila melanogaster.
Bmc Struct.Biol., 14, 2014
|
|
