2DUT
 
 | |
2H4Q
 
 | |
5FMW
 
 | | The poly-C9 component of the Complement Membrane Attack Complex | | Descriptor: | POLYC9 | | Authors: | Dudkina, N.V, Spicer, B.A, Reboul, C.F, Conroy, P.J, Lukoyanova, N, Elmlund, H, Law, R.H.P, Ekkel, S.M, Kondos, S.C, Goode, R.J.A, Ramm, G, Whisstock, J.C, Saibil, H.R, Dunstone, M.A. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the Poly-C9 Component of the Complement Membrane Attack Complex
Nat.Commun., 7, 2016
|
|
1WZ9
 
 | | The 2.1 A structure of a tumour suppressing serpin | | Descriptor: | Maspin precursor, SULFATE ION | | Authors: | Law, R.H, Irving, J.A, Buckle, A.M, Ruzyla, K, Buzza, M, Bashtannyk-Puhalovich, T.A, Beddoe, T.C, Kim, N, Worrall, D.M, Bottomley, S.P, Bird, P.I, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2005-03-03 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
1MTP
 
 | | The X-ray crystal structure of a serpin from a thermophilic prokaryote | | Descriptor: | Serine Proteinase Inhibitor (SERPIN), Chain A, Chain B | | Authors: | Irving, J.A, Cabrita, L.D, Rossjohn, J, Pike, R.N, Bottomley, S.P, Whisstock, J.C. | | Deposit date: | 2002-09-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A crystal structure of a prokaryote serpin: controlling conformational change in a heated environment
Structure, 11, 2003
|
|
4Y1S
 
 | | Structural basis for Ca2+-mediated interaction of the perforin C2 domain with lipid membranes | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Conroy, P.J, Yagi, H, Whisstock, J.C, Norton, R.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Structural Basis for Ca2+-mediated Interaction of the Perforin C2 Domain with Lipid Membranes.
J.Biol.Chem., 290, 2015
|
|
2QP2
 
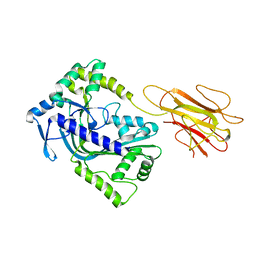 | | Structure of a MACPF/perforin-like protein | | Descriptor: | CALCIUM ION, Unknown protein | | Authors: | Rosado, C.J, Buckle, A.M, Law, R.H.P, Butcher, R.E, Kan, W.T, Bird, C.H, Ung, K, Browne, K.A, Baran, K, Bashtannyk-Puhalovich, T.A, Faux, N.G, Wong, W, Porter, C.J, Pike, R.N, Ellisdon, A.M, Pearce, M.C, Bottomley, S.P, Emsley, J, Smith, A.I, Rossjohn, J, Hartland, E.L, Voskoboinik, I, Trapani, J.A, Bird, P.I, Dunstone, M.A, Whisstock, J.C. | | Deposit date: | 2007-07-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A common fold mediates vertebrate defense and bacterial attack
Science, 317, 2007
|
|
3UB1
 
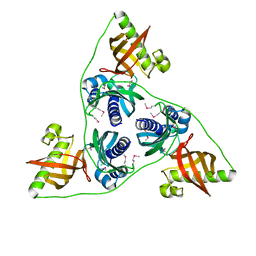 | | Ntf2 like protein involved in plasmid conjugation | | Descriptor: | DI(HYDROXYETHYL)ETHER, ORF13-like protein | | Authors: | Porter, C.J, Rosado, C.J, Bantwal, R, Bannam, T.L, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-08 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria.
Mol.Microbiol., 83, 2012
|
|
3TI7
 
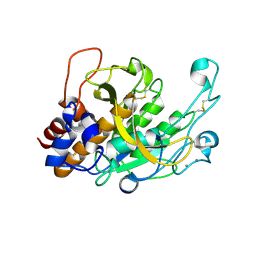 | |
2R9Y
 
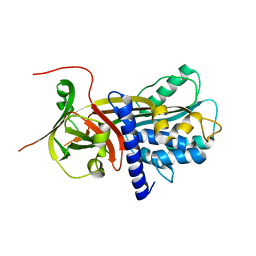 | | Structure of antiplasmin | | Descriptor: | Alpha-2-antiplasmin | | Authors: | Law, R.H.P, Sofian, T, Kan, W.T, Horvath, A.J, Hitchen, C.R, Langendorf, C.G, Buckle, A.M, Whisstock, J.C, Coughlin, P.B. | | Deposit date: | 2007-09-14 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray crystal structure of the fibrinolysis inhibitor {alpha}2-antiplasmin
Blood, 111, 2008
|
|
4F88
 
 | | X-ray Crystal Structure of PlyC | | Descriptor: | PlyCA, PlyCB | | Authors: | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F87
 
 | | X-ray Crystal Structure of PlyCB | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3KQZ
 
 | | Structure of a protease 2 | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, NONAETHYLENE GLYCOL, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KQX
 
 | | Structure of a protease 1 | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, NONAETHYLENE GLYCOL, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KR4
 
 | | Structure of a protease 3 | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KR5
 
 | | Structure of a protease 4 | | Descriptor: | (2S)-3-[(R)-[(1S)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]-2-benzylpropanoic acid, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4WVM
 
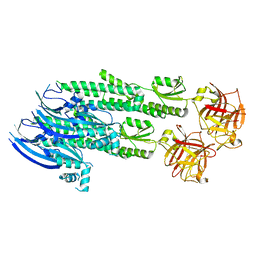 | | Stonustoxin structure | | Descriptor: | Stonustoxin subunit alpha, Stonustoxin subunit beta | | Authors: | Ellisdon, A.M, Panjikar, S, Whisstock, J.C, McGowan, S. | | Deposit date: | 2014-11-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Stonefish toxin defines an ancient branch of the perforin-like superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5HN1
 
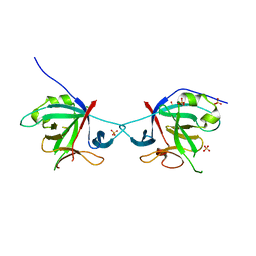 | | Crystal structure of Interleukin-37 | | Descriptor: | Interleukin-37, SULFATE ION | | Authors: | Ellisdon, A.M, Nold-Petry, C.A, Nold, M.F, Whisstock, J.C. | | Deposit date: | 2016-01-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Homodimerization attenuates the anti-inflammatory activity of interleukin-37.
Sci Immunol, 2, 2017
|
|
2VH4
 
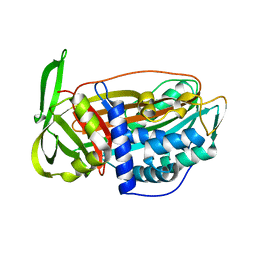 | | Structure of a loop C-sheet serpin polymer | | Descriptor: | TENGPIN | | Authors: | Zhang, Q, Law, R.H.P, Bottomley, S.P, Whisstock, J.C, Buckle, A.M. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Structural Basis for Loop C-Sheet Polymerization in Serpins.
J.Mol.Biol., 376, 2008
|
|
1SNG
 
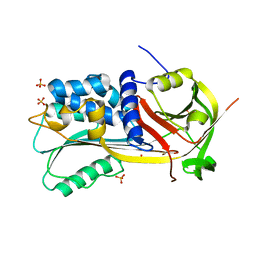 | | Structure of a Thermophilic Serpin in the Native State | | Descriptor: | COG4826: Serine protease inhibitor, SULFATE ION | | Authors: | Fulton, K.F, Buckle, A.M, Cabrita, L.D, Irving, J.A, Butcher, R.E, Smith, I, Reeve, S, Lesk, A.M, Bottomley, S.P, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2004-03-10 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition.
J.Biol.Chem., 280, 2005
|
|
1MI5
 
 | | The crystal structure of LC13 TcR in complex with HLAB8-EBV peptide complex | | Descriptor: | Epstein Barr Virus peptide, MHC heavy chain, TcR alpha chain, ... | | Authors: | Kjer-Nielsen, L, Clements, C.S, Purcell, A.W, Brooks, A.G, Whisstock, J.C, Burrows, S.R, McCluskey, J, Rossjohn, J. | | Deposit date: | 2002-08-21 | | Release date: | 2003-02-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Structural Basis for the Selection of Dominant alphabeta T Cell Receptors in Antiviral Immunity
IMMUNITY, 18, 2003
|
|
4Y1T
 
 | | Structural basis for Ca2+-mediated interaction of the perforin C2 domain with lipid membranes | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Conroy, P.J, Yagi, H, Whisstock, J.C, Norton, R.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Structural Basis for Ca2+-mediated Interaction of the Perforin C2 Domain with Lipid Membranes.
J.Biol.Chem., 290, 2015
|
|
1YXA
 
 | | Serpina3n, a murine orthologue of human antichymotrypsin | | Descriptor: | serine (or cysteine) proteinase inhibitor, clade A, member 3N | | Authors: | Horvath, A.J, Irving, J.A, Law, R.H, Rossjohn, J, Bottomley, S.P, Quinsey, N.S, Pike, R.N, Coughlin, P.B, Whisstock, J.C. | | Deposit date: | 2005-02-20 | | Release date: | 2005-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins.
J.Biol.Chem., 280, 2005
|
|
4V3A
 
 | | Membrane bound pleurotolysin prepore (TMH1 lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, CaradocDavies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
