8H8F
 
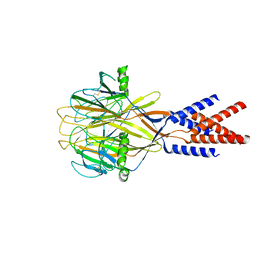 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state)
To Be Published
|
|
8H8E
 
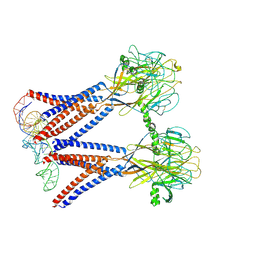 | | Structure of the dimeric Xenopus tropical acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (closed state) | | Descriptor: | Proton-activated chloride channel, tRNA (75-MER)of Spodoptera frugiperda | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the dimeric Xenopus tropical acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (closed state)
To Be Published
|
|
7WYG
 
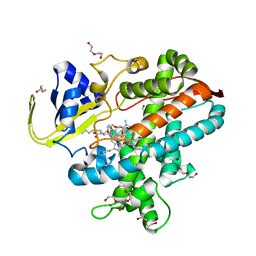 | | Crystal structure of P450BSbeta-L78I/Q85H/G290I variant in complex with palmitic acid. | | Descriptor: | Cytochrome P450 152A1, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Li, F, He, C, Wang, X. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biocatalytic Enantioselective beta-Hydroxylation of Unactivated C-H Bonds in Aliphatic Carboxylic Acids.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8A2F
 
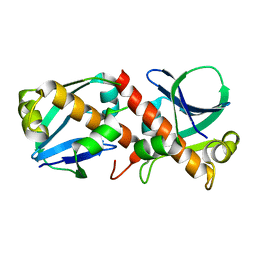 | | Crystal Structure of Ljunganvirus 1 2A protein | | Descriptor: | 2A protein | | Authors: | von Castelmur, E, Zhu, L, Wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family
To Be Published
|
|
8A2E
 
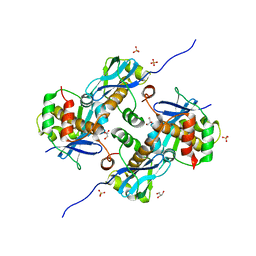 | | Crystal Structure of Human Parechovirus 3 2A protein | | Descriptor: | 2A protein, GLYCEROL, SULFATE ION | | Authors: | von Castelmur, E, Zhu, L, wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family.
To Be Published
|
|
8C15
 
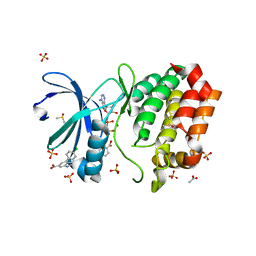 | | Aurora A kinase in complex with TPX2-inhibitor 3 | | Descriptor: | (1~{R},2~{R})-cyclohexane-1,2-dicarboxylic acid, 4-(4-chlorophenyl)-1~{H}-indole-6-carboxylic acid, ACETATE ION, ... | | Authors: | Fischer, G, Rocaboy, M, Blaszczyk, B, Moschetti, T, Wang, X, Scott, D.E, Coyne, A.G, Dagostin, C, Rooney, T, Bayly, A, Feng, J, Asteian, A, Alcaide-Lopez, A, Stockwell, S, Skidmore, J, Venkitaraman, A.R, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Selective inhibitors of the Aurora A-TPX2 protein-protein interaction exhibit in vivo efficacy as targeted anti-mitotic agents
To be published
|
|
7VUX
 
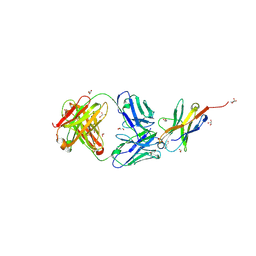 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7X25
 
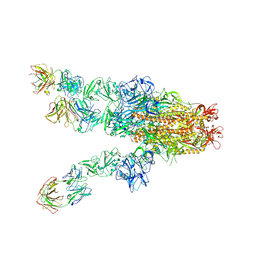 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class4 (2u1d RBD with 3Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J, Zhang, S, Zhou, H, Wang, X. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7YCX
 
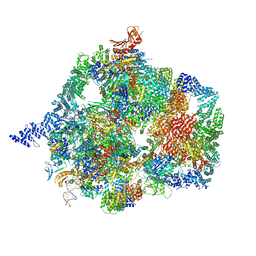 | | The structure of INTAC-PEC complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1,DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Zheng, H, Jin, Q, Wang, X, Qi, Y, Liu, W, Ren, Y, Zhao, D, Chen, F.X, Cheng, J, Chen, X, Xu, Y. | | Deposit date: | 2022-07-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural basis of INTAC-regulated transcription.
Protein Cell, 14, 2023
|
|
7YKS
 
 | |
7YKR
 
 | |
7X7R
 
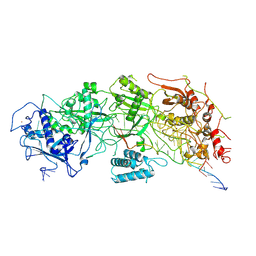 | | Cryo-EM structure of a bacterial protein | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*AP*GP*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X7A
 
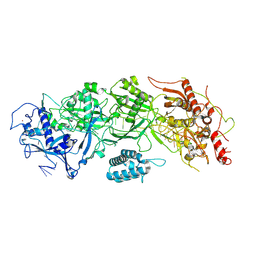 | | Cryo-EM structure of SbCas7-11 in complex with crRNA and target RNA | | Descriptor: | RAMP superfamily protein, RNA (33-MER), ZINC ION | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X8A
 
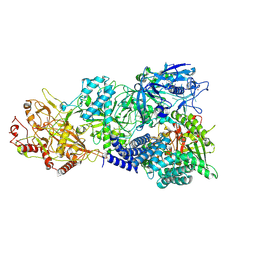 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7XC7
 
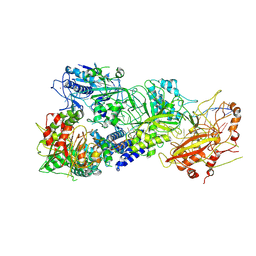 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7Y9C
 
 | |
7YF2
 
 | |
7YF4
 
 | |
7YF3
 
 | |
6JHR
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F6 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6J11
 
 | | MERS-CoV spike N-terminal domain and 7D10 scFv complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-terminal domain of Spike glycoprotein, ... | | Authors: | Zhou, H, Zhang, S, Zhang, S, Tang, W, Wang, X. | | Deposit date: | 2018-12-27 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein.
Nat Commun, 10, 2019
|
|
6JHT
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F9 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHQ
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F4 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
