4ZMH
 
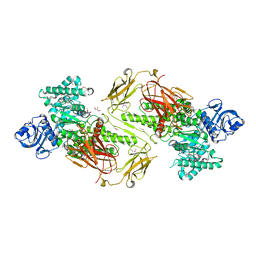 | | Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Nocek, B, Cui, H, Wang, W, Savchenko, A. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Biochemical and Structural Characterization of a Five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T.
J.Biol.Chem., 291, 2016
|
|
7L31
 
 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, strychnine bound state, 3.8 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-2, Glycine receptor subunit beta,Green fluorescent protein, ... | | Authors: | Yu, H, Wang, W. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
7KUY
 
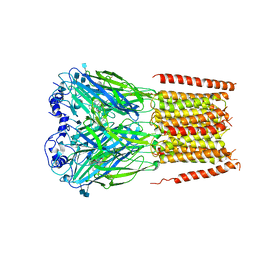 | |
7VIB
 
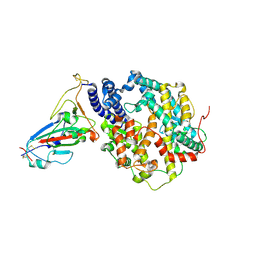 | | Crystal structure of human ACE2 and GX/P2V RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein, ZINC ION | | Authors: | Guo, Y, Cao, W, Jia, N, Wang, W, Yuan, S, Wang, Y. | | Deposit date: | 2021-09-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human ACE2 and GX/P2V RBD
To Be Published
|
|
5Y51
 
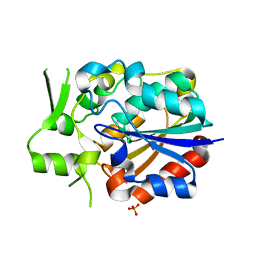 | | Crystal structure of PytH_H230A | | Descriptor: | Pyrethroid hydrolase, SULFATE ION | | Authors: | Xu, D.Q, Ran, T.T, He, J, Wang, W.W. | | Deposit date: | 2017-08-06 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Catalytic Mechanism of a Novel Pyrethroid Hydrolase from Sphingobium faniae JZ-2
To Be Published
|
|
7E1V
 
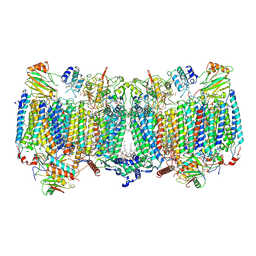 | | Cryo-EM structure of apo hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, CARDIOLIPIN, ... | | Authors: | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-13 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
7E1W
 
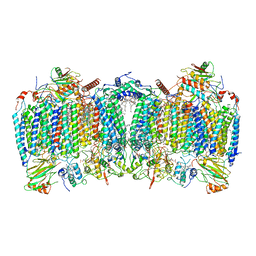 | | Cryo-EM structure of hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV in the presence of Q203 | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, 6-chloranyl-2-ethyl-N-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperidin-1-yl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide, ... | | Authors: | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-27 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
7E1X
 
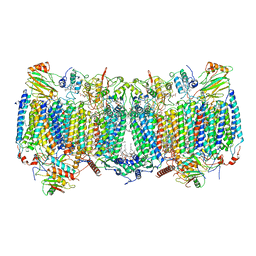 | | Cryo-EM structure of hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV in presence of TB47 | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, 5-methoxy-2-methyl-~{N}-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperidin-1-yl]phenyl]methyl]pyrazolo[1,5-a]pyridine-3-carboxamide, ... | | Authors: | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-27 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
5Y0E
 
 | |
5Y5V
 
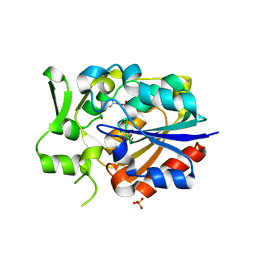 | |
4PPM
 
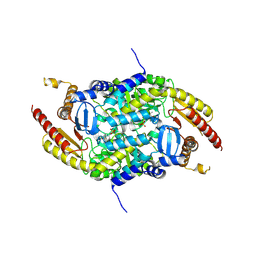 | | Crystal structure of PigE: a transaminase involved in the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP) from Serratia sp. FS14 | | Descriptor: | Aminotransferase, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lou, X.D, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2014-02-27 | | Release date: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of PigE: a transaminase involved in the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP) from Serratia sp. FS14
Biochem.Biophys.Res.Commun., 447, 2014
|
|
5BKG
 
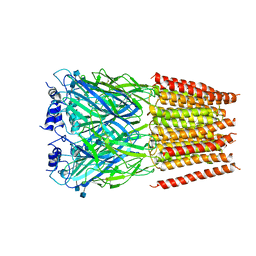 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, glycine bound, (semi)open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alpha-2, ... | | Authors: | Yu, H, Wang, W. | | Deposit date: | 2021-03-19 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
5BKF
 
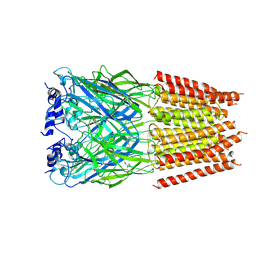 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, Glycine bound, desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alpha-2, ... | | Authors: | Yu, H, Wang, W. | | Deposit date: | 2021-03-19 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
5CIO
 
 | | Crystal structure of PqqF | | Descriptor: | ZINC ION, pyrroloquinoline quinone biosynthesis protein PqqF | | Authors: | Wei, Q, Xu, D, Ran, T, Wang, W. | | Deposit date: | 2015-07-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway
J.Biol.Chem., 291, 2016
|
|
5CZW
 
 | | Crystal structure of myroilysin | | Descriptor: | Myroilysin, ZINC ION | | Authors: | Zhou, J, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2015-08-01 | | Release date: | 2016-08-03 | | Last modified: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Myroilysin is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Activated by a Cysteine-switch Mechanism
J. Biol. Chem., 292, 2017
|
|
5VFS
 
 | | Nucleotide-Driven Triple-State Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFT
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFO
 
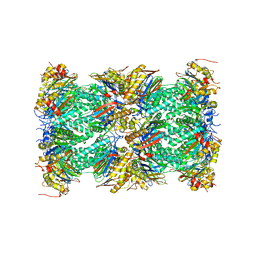 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-08 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFQ
 
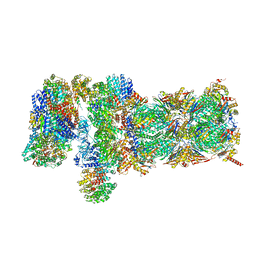 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5WBS
 
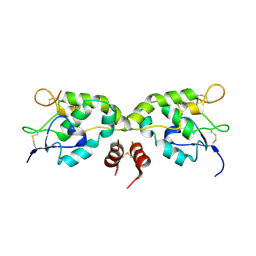 | | Crystal structure of Frizzled-7 CRD with an inhibitor peptide Fz7-21 | | Descriptor: | Frizzled-7,inhibitor peptide Fz7-21 | | Authors: | Nile, A.H, Mukund, S, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
5WUW
 
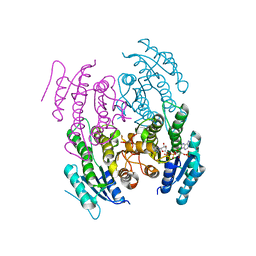 | |
5WUL
 
 | |
5FHP
 
 | | SeMet regulator of nicotine degradation | | Descriptor: | GLYCEROL, MALONIC ACID, NicR | | Authors: | Zhang, K, Tang, H, Wu, G, Wang, W, Hu, H, Xu, P. | | Deposit date: | 2015-12-22 | | Release date: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Co-crystal Structure of NicR2_Hsp
To Be Published
|
|
4O14
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1C
 
 | | The crystal structures of a mutant NAMPT H191R | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
