4O10
 
 | | Structural and Biochemical Analyses of the Catalysis and Potency Impact of Inhibitor Phosphoribosylation by Human Nicotinamide Phosphoribosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}indolizine-7-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Wang, W. | | Deposit date: | 2013-12-14 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
4O28
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-17 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1D
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
5AD2
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
5AD3
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
3ET2
 
 | |
3ET0
 
 | |
3ET1
 
 | |
3ET3
 
 | |
6JFB
 
 | |
4QP1
 
 | |
4QP9
 
 | |
4QP3
 
 | |
4QP4
 
 | |
4QPA
 
 | |
4QP7
 
 | |
4QP6
 
 | |
4QP2
 
 | |
4QP8
 
 | |
5VFP
 
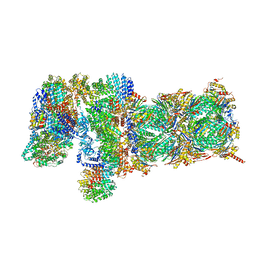 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFR
 
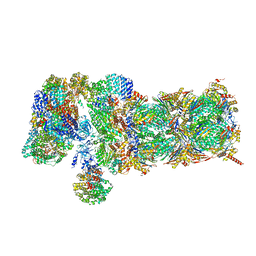 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFU
 
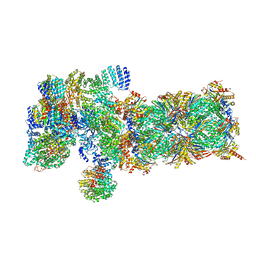 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFS
 
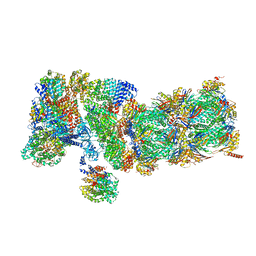 | | Nucleotide-Driven Triple-State Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFT
 
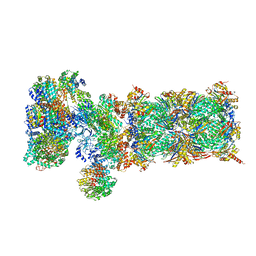 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFO
 
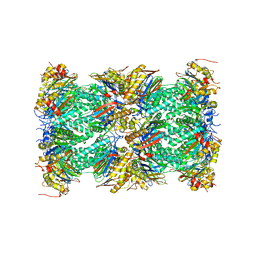 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-08 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
