1QGM
 
 | |
1CE4
 
 | |
1IEH
 
 | | SOLUTION STRUCTURE OF A SOLUBLE SINGLE-DOMAIN ANTIBODY WITH HYDROPHOBIC RESIDUES TYPICAL OF A VL/VH INTERFACE | | Descriptor: | BRUC.D4.4 | | Authors: | Vranken, W, Tolkatchev, D, Xu, P, Tanha, J, Chen, Z, Narang, S, Ni, F. | | Deposit date: | 2001-04-09 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a llama single-domain antibody with hydrophobic residues typical of the VH/VL interface.
Biochemistry, 41, 2002
|
|
1I8Y
 
 | |
1I8X
 
 | |
3ZPD
 
 | |
6QD6
 
 | | Molecular scaffolds expand the nanobody toolkit for cryo-EM applications: crystal structure of Mb-cHopQ-Nb207 | | Descriptor: | CHLORIDE ION, Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207 | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkonig, A, Zogg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-18 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM
Nat.Methods, 18, 2021
|
|
6QFA
 
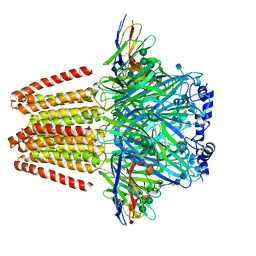 | | CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit beta-3, HISTAMINE, ... | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkoening, A, Zoegg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2019-01-09 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
1G26
 
 | |
3ZBE
 
 | |
