2KMY
 
 | |
2KSU
 
 | |
2MXL
 
 | |
2MXK
 
 | |
2MXJ
 
 | |
2MS3
 
 | |
2LGN
 
 | | Lactococcin 972 | | Descriptor: | Lactococcin 972 | | Authors: | Turner, D.L, Lamosa, P, Martinez, B. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-01 | | Method: | SOLUTION NMR | | Cite: | Structure and properties of lactococcin 972 from Lactococcus lactis
To be Published
|
|
1A2I
 
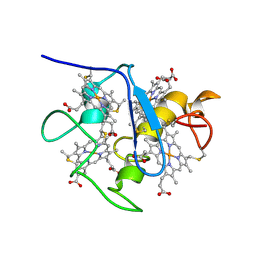 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Kastrau, D.H.K, Costa, H.S, Legall, J, Turner, D.L, Santos, H, Xavier, A.V. | | Deposit date: | 1998-01-05 | | Release date: | 1998-07-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Desulfovibrio vulgaris (Hildenborough) ferrocytochrome c3: structural basis for functional cooperativity.
J.Mol.Biol., 281, 1998
|
|
7RQ5
 
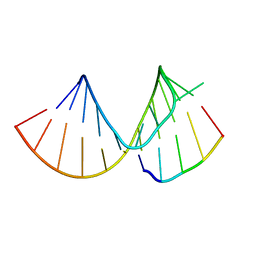 | |
1E8E
 
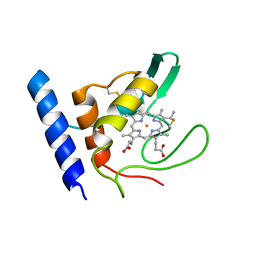 | | Solution Structure of Methylophilus methylotrophus Cytochrome c''. Insights into the Structural Basis of Haem-Ligand Detachment | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Brennan, L, Turner, D.L, Fareleira, P, Santos, H. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-20 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methylophilus Methylotrophus Cytochrome C": Insights Into the Structural Basis of Haem-Ligand Detachment
J.Mol.Biol., 308, 2001
|
|
2BPN
 
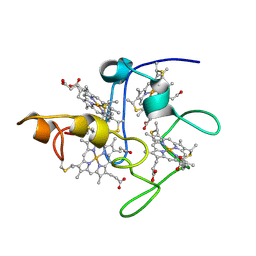 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERRICYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Aguiar, A.P, Brennan, L, Xavier, A.V, Turner, D.L. | | Deposit date: | 2005-04-21 | | Release date: | 2006-03-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Tetrahaem Ferricytochrome C(3) from Desulfovibrio Vulgaris (Hildenborough) and its K45Q Mutant: The Molecular Basis of Cooperativity.
Biochim.Biophys.Acta, 1757, 2006
|
|
333D
 
 | | THE CRYSTAL STRUCTURE OF AN RNA OLIGOMER INCORPORATING TANDEM ADENOSINE-INOSINE MISMATCHES | | Descriptor: | CALCIUM ION, RNA (5'-R(*CP*GP*CP*AP*IP*GP*CP*G)-3') | | Authors: | Carter, R.J, Baeyens, K.J, SantaLucia Jr, J, Turner, D.H, Holbrook, S.R. | | Deposit date: | 1997-05-20 | | Release date: | 1997-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The crystal structure of an RNA oligomer incorporating tandem adenosine-inosine mismatches.
Nucleic Acids Res., 25, 1997
|
|
1QN0
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Teodoro, M.L, Brennan, L, Legall, J, Santos, H, Xavier, A.V, Turner, D.L. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochrome C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1QN1
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
6N8H
 
 | |
1GK5
 
 | | Solution Structure the mEGF/TGFalpha44-50 chimeric growth factor | | Descriptor: | Pro-epidermal growth factor,Protransforming growth factor alpha | | Authors: | Chamberlin, S.G, Brennan, L, Puddicombe, S.M, Davies, D.E, Turner, D.L. | | Deposit date: | 2001-08-08 | | Release date: | 2002-08-08 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Megf/Tgfalpha44-50 Chimeric Growth Factor.
Eur.J.Biochem., 268, 2001
|
|
1MIS
 
 | |
5NEV
 
 | | CDK2/Cyclin A in complex with compound 73 | | Descriptor: | 4-[[6-(3-phenylphenyl)-7~{H}-purin-2-yl]amino]benzenesulfonamide, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E.M, Newell, D.R, Turner, D, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Cano, G. | | Deposit date: | 2017-03-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
1TUT
 
 | | J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns | | Descriptor: | 5'-R(*GP*AP*GP*GP*AP*AP*GP*GP*CP*GP*A)-3', 5'-R(*UP*CP*GP*UP*UP*AP*AP*UP*CP*UP*C)-3' | | Authors: | Znosko, B.M, Kennedy, S.D, Wille, P.C, Krugh, T.R, Turner, D.H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-12-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural Features and Thermodynamics of the J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns.
Biochemistry, 43, 2004
|
|
1NTT
 
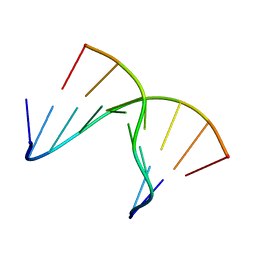 | | 5'(dCPCPUPCPCPUPUP)3':(rAGGAGGAAA)5', where P=propynyl | | Descriptor: | 5'-D(*CP*(5PC)P*(PDU)P*(5PC)P*(5PC)P*(PDU)P*(PDU))-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2021-07-28 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
1NTQ
 
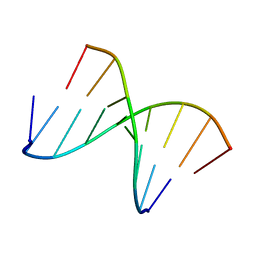 | | 5'(dCCUCCUU)3':3'(rAGGAGGAAA)5' | | Descriptor: | 5'-D(*CP*CP*UP*CP*CP*UP*U)-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
1NTS
 
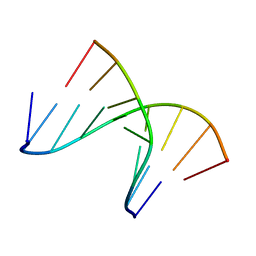 | | 5'(dCCPUPCPCPUPUP)3':3'(rAGGAGGAAA)5', where P=propynyl | | Descriptor: | 5'-D(*(5PC)P*(5PC)P*(PDU)P*(5PC)P*(5PC)P*(PDU)P*(PDU))-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2021-07-28 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
1XV0
 
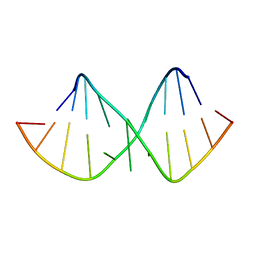 | | Solution NMR structure of RNA internal loop with three consecutive sheared GA pairs in 5'GGUGGAGGCU/3'PCCGAAGCCG | | Descriptor: | 5'-R(*GP*CP*CP*GP*AP*AP*GP*CP*CP*(P5P)-3', 5'-R(*GP*GP*UP*GP*GP*AP*GP*GP*CP*U)-3' | | Authors: | Chen, G, Znosko, B.M, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an RNA Internal Loop with Three Consecutive Sheared GA Pairs
Biochemistry, 44, 2005
|
|
6N8I
 
 | |
6N8F
 
 | |
