1NXB
 
 | |
1ANV
 
 | | ADENOVIRUS 5 DBP/URANYL FLUORIDE SOAK | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, URANYL (VI) ION, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1996-03-14 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change of the adenovirus DNA-binding protein induced by soaking crystals with K3UO2F5 solutions.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4RVE
 
 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*GP*GP*GP*AP*TP*AP*TP*CP*CP*C)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilso, K.S. | | Deposit date: | 1993-02-18 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
1ADV
 
 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1ADU
 
 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1A87
 
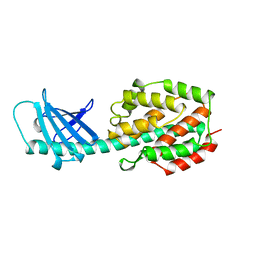 | | COLICIN N | | Descriptor: | COLICIN N | | Authors: | Vetter, I.R, Parker, M.W, Tucker, A.D, Lakey, J.H, Pattus, F, Tsernoglou, D. | | Deposit date: | 1998-04-03 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a colicin N fragment suggests a model for toxicity.
Structure, 6, 1998
|
|
1COL
 
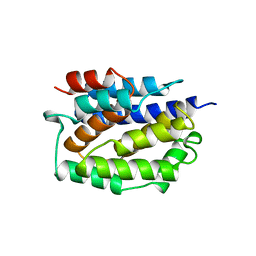 | | REFINED STRUCTURE OF THE PORE-FORMING DOMAIN OF COLICIN A AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | COLICIN A | | Authors: | Parker, M.W, Postma, J.P.M, Pattus, F, Tucker, A.D, Tsernoglou, D. | | Deposit date: | 1991-07-06 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined structure of the pore-forming domain of colicin A at 2.4 A resolution.
J.Mol.Biol., 224, 1992
|
|
1GJY
 
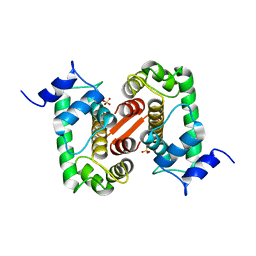 | | The X-ray structure of the Sorcin Calcium Binding Domain (SCBD) provides insight into the phosphorylation and calcium dependent processess | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Ilari, A, Johnson, K.A, Nastopoulos, V, Tsernoglou, D, Chiancone, E. | | Deposit date: | 2001-08-06 | | Release date: | 2002-04-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Sorcin Calcium Binding Domain Provides a Model of Ca(2+)-Dependent Processes in the Full-Length Protein
J.Mol.Biol., 317, 2002
|
|
1HG8
 
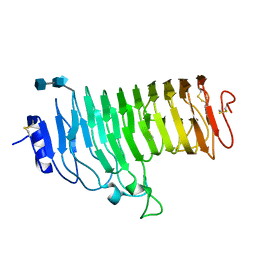 | | Endopolygalacturonase from the phytopathogenic fungus Fusarium moniliforme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOPOLYGALACTURONASE | | Authors: | Federici, L, Caprari, C, Mattei, B, Savino, C, De Lorenzo, G, Cervone, F, Tsernoglou, D. | | Deposit date: | 2000-12-13 | | Release date: | 2001-11-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Requirements of Endopolygalacturonase for the Interaction with Pgip (Polygalacturonase-Inhibiting Protein)
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2RVE
 
 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*CP*TP*CP*G)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilson, K.S. | | Deposit date: | 1991-03-19 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
1XG2
 
 | | Crystal structure of the complex between pectin methylesterase and its inhibitor protein | | Descriptor: | Pectinesterase 1, Pectinesterase inhibitor | | Authors: | Di Matteo, A, Raiola, A, Camardella, L, Giovane, A, Bonivento, D, De Lorenzo, G, Cervone, F, Bellincampi, D, Tsernoglou, D. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Interaction between Pectin Methylesterase and a Specific Inhibitor Protein
Plant Cell, 17, 2005
|
|
1PRE
 
 | | PROAEROLYSIN | | Descriptor: | PROAEROLYSIN | | Authors: | Parker, M.W, Buckley, J.T, Postma, J.P.M, Tucker, A.D, Tsernoglou, D. | | Deposit date: | 1995-09-15 | | Release date: | 1996-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states.
Nature, 367, 1994
|
|
1ROP
 
 | |
1S5J
 
 | | Insight in DNA Replication: The crystal structure of DNA Polymerase B1 from the archaeon Sulfolobus solfataricus | | Descriptor: | DNA polymerase I, MAGNESIUM ION, SULFATE ION | | Authors: | Savino, C, Federici, L, Nastopoulos, V, Johnson, K.A, Pisani, F.M, Rossi, M, Tsernoglou, D. | | Deposit date: | 2004-01-21 | | Release date: | 2004-11-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into DNA replication: the crystal structure of DNA polymerase B1 from the archaeon Sulfolobus solfataricus
Structure, 12, 2004
|
|
1OGQ
 
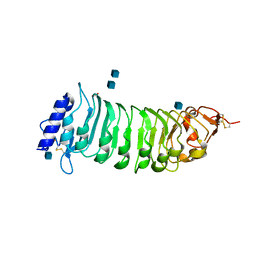 | | The crystal structure of PGIP (polygalacturonase inhibiting protein), a leucine rich repeat protein involved in plant defense | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, POLYGALACTURONASE INHIBITING PROTEIN | | Authors: | Di Matteo, A, Federici, L, Mattei, B, Salvi, G, Johnson, K.A, Savino, C, De Lorenzo, G, Tsernoglou, D, Cervone, F. | | Deposit date: | 2003-05-08 | | Release date: | 2003-07-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polygalacturonase-Inhibiting Protein (Pgip), a Leucine-Rich Repeat Protein Involved in Plant Defense
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1QKD
 
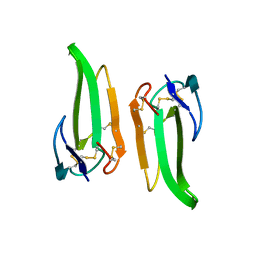 | | ERABUTOXIN | | Descriptor: | ERABUTOXIN A | | Authors: | Nastopoulos, V, Kanellopoulos, P.N, Tsernoglou, D. | | Deposit date: | 1998-01-16 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of dimeric and monomeric erabutoxin a refined at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1OE7
 
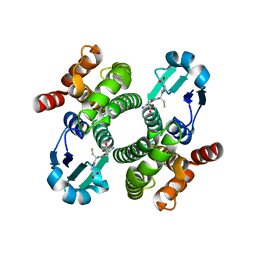 | |
1OE8
 
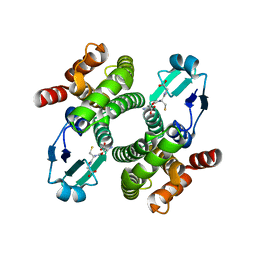 | |
1QKE
 
 | | ERABUTOXIN | | Descriptor: | ERABUTOXIN A, SULFATE ION | | Authors: | Nastopoulos, V, Kanellopoulos, P.N, Tsernoglou, D. | | Deposit date: | 1998-01-16 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of dimeric and monomeric erabutoxin a refined at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1QGH
 
 | | THE X-RAY STRUCTURE OF THE UNUSUAL DODECAMERIC FERRITIN FROM LISTERIA INNOCUA, REVEALS A NOVEL INTERSUBUNIT IRON BINDING SITE. | | Descriptor: | FE (III) ION, NON-HEME IRON-CONTAINING FERRITIN | | Authors: | Ilari, A, Stefanini, S, Chiancone, E, Tsernoglou, D. | | Deposit date: | 1999-04-27 | | Release date: | 2000-01-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site.
Nat.Struct.Biol., 7, 2000
|
|
1QI7
 
 | | THE CRYSTAL STRUCTURE AT 2.0 A OF SAPORIN SO6, A RIBOSOME INACTIVATING PROTEIN FROM SAPONARIA OFFICINALIS | | Descriptor: | PROTEIN (N-GLYCOSIDASE), SULFATE ION | | Authors: | Savino, C, Federici, L, Ippoliti, R, Lendaro, E, Tsernoglou, D. | | Deposit date: | 1999-06-08 | | Release date: | 2000-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of saporin SO6 from Saponaria officinalis and its interaction with the ribosome.
FEBS Lett., 470, 2000
|
|
