4U7W
 
 | | The crystal structure of the terminal R domain from the myxalamid PKS-NRPS biosynthetic pathway | | Descriptor: | ACETATE ION, MxaA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsai, S.C, Keasling, J.D, Luo, R, Barajas, J.F, Phelan, R.M, Schaub, A.J, Kliewer, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols.
Chem.Biol., 22, 2015
|
|
4W4T
 
 | | The crystal structure of the terminal R domain from the myxalamid PKS-NRPS biosynthetic pathway | | Descriptor: | ACETATE ION, MxaA | | Authors: | Tsai, S.C, Keasling, J.D, Luo, R, Barajas, J.F, Phelan, R.M, Schaub, A.J, Kliewer, J. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols.
Chem.Biol., 22, 2015
|
|
4WV3
 
 | | Crystal structure of the anthranilate CoA ligase AuaEII in complex with anthranoyl-AMP | | Descriptor: | 5'-O-[(S)-[(2-aminobenzoyl)oxy](hydroxy)phosphoryl]adenosine, Anthranilate-CoA ligase | | Authors: | Tsai, S.C, Muller, R, Jackson, D.R, Tu, S.S, Nguyen, M, Barajas, J.F, Pistorious, D, Krug, D. | | Deposit date: | 2014-11-04 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structural Insights into Anthranilate Priming during Type II Polyketide Biosynthesis.
Acs Chem.Biol., 11, 2016
|
|
4XRW
 
 | | Crystal structure of the di-domain ARO/CYC BexL from the BE-7585A biosynthetic pathway | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BexL | | Authors: | Tsai, S.C, Caldara-Festin, G.M, Jackson, D.R, Aguilar, S, Patel, A, Nguyen, M, Sasaki, E, Valentic, T.R, Barajas, J.F, Vo, M, Khanna, A, Liu, H.-W. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional analysis of two di-domain aromatase/cyclases from type II polyketide synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XRT
 
 | | Crystal structure of the di-domain ARO/CYC StfQ from the steffimycin biosynthetic pathway | | Descriptor: | FORMIC ACID, StfQ Aromatase/Cyclase | | Authors: | Tsai, S.C, Caldara-Festin, G.M, Jackson, D.R, Aguilar, S, Patel, A, Nguyen, M, Sasaki, E, Valentic, T.R, Barajas, J.F, Vo, M, Khanna, A, Liu, H.-W. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural and functional analysis of two di-domain aromatase/cyclases from type II polyketide synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X4J
 
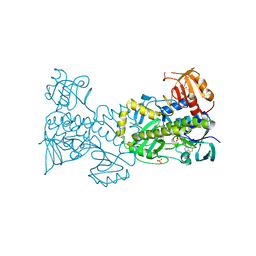 | | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative oxygenase, SULFATE ION | | Authors: | Tsai, S.-C, Jackson, D.R, Patel, A, Barajas, J.F, Rohr, J, Yu, X, Liu, H.-W, Sasaki, E, Calveras, J, Metsa-Ketela, M. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis
To Be Published
|
|
5KBZ
 
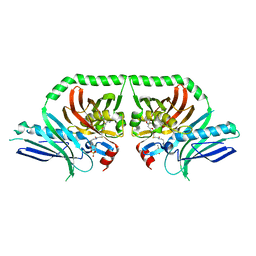 | | Structure of the PksA Product Template domain in complex with a phosphopantetheine mimetic | | Descriptor: | (14R)-14-hydroxy-15,15-dimethyl-1-[5-({[(5-methyl-1,2-oxazol-3-yl)methyl]sulfanyl}methyl)-1,2-oxazol-3-yl]-4,9,13-trioxo-2-thia-5,8,12-triazahexadecan-16-yl dihydrogen phosphate, Noranthrone synthase | | Authors: | Tsai, S.C, Burkart, M.D, Townsend, C.A, Barajas, J.F, Shakya, G, Topper, C.L, Moreno, G, Jackson, D.R, Rivera, H, La Clair, J.J, Vagstad, A. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Polyketide mimetics yield structural and mechanistic insights into product template domain function in nonreducing polyketide synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3H2X
 
 | |
1MNA
 
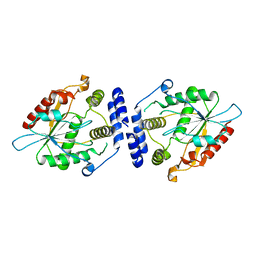 | | Thioesterase Domain of Picromycin Polyketide Synthase (PICS TE), pH 8.0 | | Descriptor: | polyketide synthase IV | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
1MNQ
 
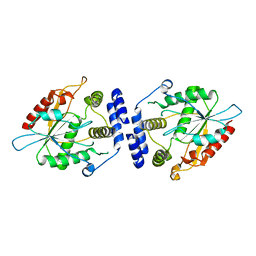 | | Thioesterase Domain of Picromycin Polyketide Synthase (PICS TE), pH 8.4 | | Descriptor: | polyketide synthase IV | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
1MO2
 
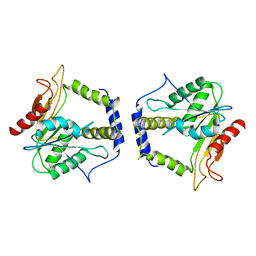 | | Thioesterase Domain from 6-Deoxyerythronolide Synthase (DEBS TE), pH 8.5 | | Descriptor: | Erythronolide synthase, modules 5 and 6 | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
1MN6
 
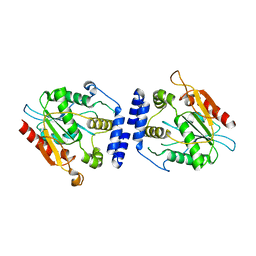 | | Thioesterase Domain from Picromycin Polyketide Synthase, pH 7.6 | | Descriptor: | polyketide synthase IV | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
3BB8
 
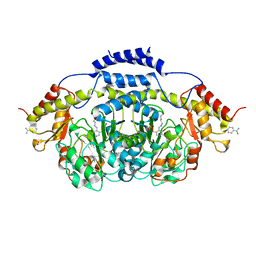 | | E1 Dehydrase H220K Mutant | | Descriptor: | BENZAMIDINE, CDP-4-keto-6-deoxy-D-glucose-3-dehydrase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tsai, S.-C, Smith, P. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | E1 Dehydrase H220K Mutant
To be Published
|
|
3BCX
 
 | | E1 Dehydrase | | Descriptor: | BENZAMIDINE, CDP-6-deoxy-L-threo-D-glycero-4-hexulose-3-dehydrase, FE (III) ION | | Authors: | Tsai, S.-C, Smith, P. | | Deposit date: | 2007-11-13 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E1 Dehydrase.
To be Published
|
|
1KEZ
 
 | | Crystal Structure of the Macrocycle-forming Thioesterase Domain of Erythromycin Polyketide Synthase (DEBS TE) | | Descriptor: | ERYTHRONOLIDE SYNTHASE | | Authors: | Tsai, S.-C, Miercke, L.J.W, Krucinski, J, Gokhale, R, Chen, J.C.-H, Foster, P.G, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2001-11-19 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5MTI
 
 | | Bamb_5917 Acyl-Carrier Protein | | Descriptor: | Phosphopantetheine-binding protein | | Authors: | Gallo, A, Kosol, S, Griffiths, D, Masschelein, J, Alkhalaf, L, Smith, H, Valentic, T, Tsai, S, Challis, G, Lewandowski, J.R. | | Deposit date: | 2017-01-09 | | Release date: | 2018-08-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase
Nat.Chem., 2019
|
|
4XSA
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
3B6Z
 
 | | Lovastatin polyketide enoyl reductase (LovC) complexed with 2'-phosphoadenosyl isomer of crotonoyl-CoA | | Descriptor: | Enoyl reductase, GLYCEROL, S-{(9R,13R,15S)-17-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-9,13,15-trihydroxy-10,10-dimethyl-13,15-dioxido-4,8-dioxo-12,14,16-trioxa-3,7-diaza-13,15-diphosphaheptadec-1-yl}(2E)-but-2-enethioate | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B70
 
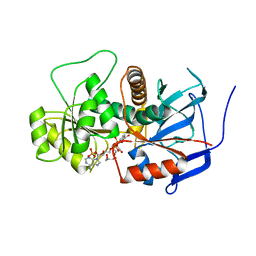 | | Crystal structure of Aspergillus terreus trans-acting lovastatin polyketide enoyl reductase (LovC) with bound NADP | | Descriptor: | Enoyl reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DC0
 
 | |
4DBZ
 
 | |
4DC1
 
 | |
5TII
 
 | |
5THQ
 
 | |
5INF
 
 | | Structural basis for acyl-CoA carboxylase-mediated assembly of unusual polyketide synthase extender units incorporated into the stambomycin antibiotics | | Descriptor: | Carboxyl transferase, HEXANOYL-COENZYME A | | Authors: | Valentic, T.R, Ray, L, Miyazawa, T, Withall, D.M, Song, L, Osada, H, Tsai, S.C, Challis, G.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units.
Nat Commun, 7, 2016
|
|
