1DWU
 
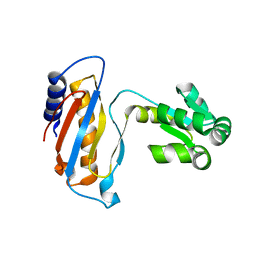 | | Ribosomal protein L1 | | Descriptor: | RIBOSOMAL PROTEIN L1 | | Authors: | Tishchenko, S.V, Nevskaya, N.A, Pavelyev, M.N, Nikonov, S.V, Garber, M.B, Piendl, W. | | Deposit date: | 1999-12-13 | | Release date: | 2000-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Ribosomal Protein L1 from Methanococcus Thermolithotrophicus. Functionally Important Structural Invariants on the L1 Surface
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3QOY
 
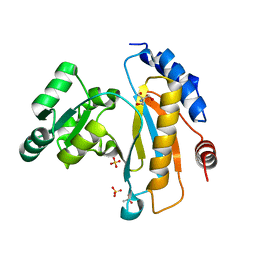 | | Crystal structure of ribosomal protein L1 from Aquifex aeolicus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 50S ribosomal protein L1, ACETIC ACID, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, S.V, Nikonova, E.U, Shkliaeva, A.A, Garber, M.B, Nikonov, S.V, Nevskaya, N.A. | | Deposit date: | 2011-02-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Ribosomal Protein L1 from the Bacterium Aquifex Aeolicus
Crystallography Reports, 56, 2011
|
|
4X9C
 
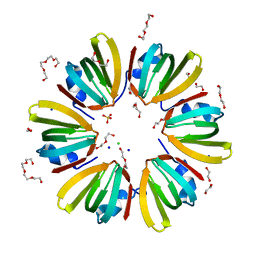 | | 1.4A crystal structure of Hfq from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, S.V, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
4F9T
 
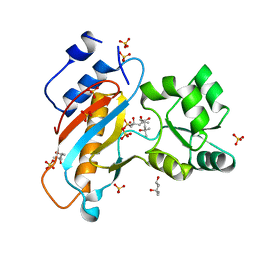 | | Ribosomal protein L1 from Thermus thermophilus with substitution Thr217Ala | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L1, ... | | Authors: | Kljashtorny, V.G, Volchkov, S.A, Nikonova, E.Y, Kostareva, O.S, Tishchenko, S.V, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | [Crystal structures of mutant ribosomal proteins L1].
MOL.BIOL.(MOSCOW), 41, 2007
|
|
4LQ4
 
 | | crystal structure of mutant ribosomal protein L1 from Methanococcus jannaschii with deletion of 8 residues from C-terminus | | Descriptor: | 50S ribosomal protein L1, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, S.V, Shkliaeva, A.A, Garber, M.B, Nikonov, S.V, Sarskikh, A.V. | | Deposit date: | 2013-07-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a mutant of archaeal ribosomal protein L1 from Methanococcus jannaschii
Crystallography Reports, 59, 2014
|
|
8P9U
 
 | | Crystal Structure of Two-Domain Laccase mutant M199A/D268N from Streptomyces griseoflavus | | Descriptor: | CALCIUM ION, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Kolyadenko, I.A, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight into the Amino Acid Environment of the Two-Domain Laccase's Trinuclear Copper Cluster.
Int J Mol Sci, 24, 2023
|
|
4REO
 
 | | Mutant ribosomal protein l1 from thermus thermophilus with threonine 217 replaced by valine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L1, GLYCINE, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Tishchenko, S.V, Nikonov, S.V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8P9V
 
 | | Crystal Structure of Two-Domain Laccase mutant M199G/R240H/D268N from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, OXYGEN MOLECULE, PEROXIDE ION, ... | | Authors: | Kolyadenko, I.A, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into the Amino Acid Environment of the Two-Domain Laccase's Trinuclear Copper Cluster.
Int J Mol Sci, 24, 2023
|
|
8B7W
 
 | | Complex IL-17A/anti-IL-17A-76 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-17A, ... | | Authors: | Kostareva, O.S, Svoeglazova, A, Kolyadenko, I.A, Dzhus, U.F, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2022-10-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two Epitope Regions Revealed in the Complex of IL-17A and Anti-IL-17A V H H Domain.
Int J Mol Sci, 23, 2022
|
|
4X9D
 
 | | High-resolution structure of Hfq from Methanococcus jannaschii in complex with UMP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, E.Y, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
5DY9
 
 | | Y68T Hfq from Methanococcus jannaschii in complex with AMP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nikulin, A.D, Mikhailina, A.O, Lekontseva, N.V, Balobanov, V.A, Nikonova, E.Y, Tishchenko, S.V. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-28 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
6T7O
 
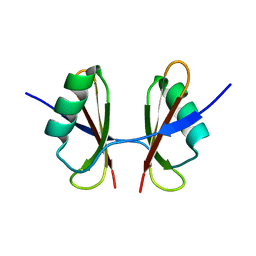 | | X-ray structure of the C-terminal domain of S. aureus Hibernating Promoting Factor (CTD-SaHPF) | | Descriptor: | Ribosome hibernation promotion factor | | Authors: | Fatkhullin, B.F, Khusainov, I.S, Ayupov, R.K, Gabdulkhakov, A.G, Tishchenko, S.V, Validov, S.Z, Yusupov, M.M. | | Deposit date: | 2019-10-22 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.60003626 Å) | | Cite: | Dimerization of long hibernation promoting factor from Staphylococcus aureus: Structural analysis and biochemical characterization.
J.Struct.Biol., 209, 2020
|
|
5MKN
 
 | |
5MKI
 
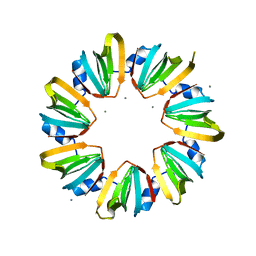 | |
5MKL
 
 | |
