5FPS
 
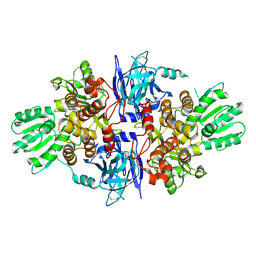 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 3-aminobenzene-1,2-dicarboxylic acid (AT1246) in an alternate binding site. | | Descriptor: | 3-AMINOBENZENE-1,2-DICARBOXYLIC ACID, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1PPT
 
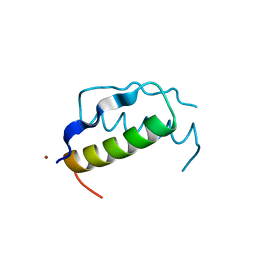 | | X-RAY ANALYSIS (1.4-ANGSTROMS RESOLUTION) OF AVIAN PANCREATIC POLYPEPTIDE. SMALL GLOBULAR PROTEIN HORMONE | | Descriptor: | AVIAN PANCREATIC POLYPEPTIDE, ZINC ION | | Authors: | Blundell, T.L, Pitts, J.E, Tickle, I.J, Wood, S.P. | | Deposit date: | 1981-01-16 | | Release date: | 1981-02-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray analysis (1. 4-A resolution) of avian pancreatic polypeptide: Small globular protein hormone.
Proc.Natl.Acad.Sci.Usa, 78, 1981
|
|
1XY2
 
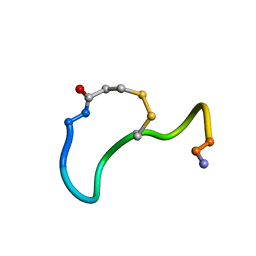 | | CRYSTAL STRUCTURE ANALYSIS OF DEAMINO-OXYTOCIN. CONFORMATIONAL FLEXIBILITY AND RECEPTOR BINDING | | Descriptor: | OXYTOCIN | | Authors: | Cooper, S, Blundell, T.L, Pitts, J.E, Wood, S.P, Tickle, I.J. | | Deposit date: | 1987-06-05 | | Release date: | 1988-04-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding.
Science, 232, 1986
|
|
1XY1
 
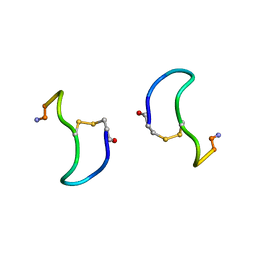 | | CRYSTAL STRUCTURE ANALYSIS OF DEAMINO-OXYTOCIN. CONFORMATIONAL FLEXIBILITY AND RECEPTOR BINDING | | Descriptor: | BETA-MERCAPTOPROPIONATE-OXYTOCIN | | Authors: | Husain, J, Blundell, T.L, Wood, S.P, Tickle, I.J, Cooper, S, Pitts, J.E. | | Deposit date: | 1987-06-05 | | Release date: | 1988-04-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding.
Science, 232, 1986
|
|
1MPP
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES. V. STRUCTURE AND REFINEMENT AT 2.0 ANGSTROMS RESOLUTION OF THE ASPARTIC PROTEINASE FROM MUCOR PUSILLUS | | Descriptor: | PEPSIN, SULFATE ION | | Authors: | Newman, M, Watson, F, Roychowdhury, P, Jones, H, Badasso, M, Cleasby, A, Wood, S.P, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1992-02-19 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of aspartic proteinases. V. Structure and refinement at 2.0 A resolution of the aspartic proteinase from Mucor pusillus.
J.Mol.Biol., 230, 1993
|
|
1MOM
 
 | | CRYSTAL STRUCTURE OF MOMORDIN, A TYPE I RIBOSOME INACTIVATING PROTEIN FROM THE SEEDS OF MOMORDICA CHARANTIA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MOMORDIN | | Authors: | Husain, J, Tickle, I.J, Wood, S.P. | | Deposit date: | 1994-03-04 | | Release date: | 1994-05-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of momordin, a type I ribosome inactivating protein from the seeds of Momordica charantia.
FEBS Lett., 342, 1994
|
|
5PEP
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEASES. II. THREE-DIMENSIONAL STRUCTURE OF THE HEXAGONAL CRYSTAL FORM OF PORCINE PEPSIN AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | PEPSIN | | Authors: | Cooper, J.B, Khan, G, Taylor, G, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1990-05-30 | | Release date: | 1990-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | X-ray analyses of aspartic proteinases. II. Three-dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3 A resolution.
J.Mol.Biol., 214, 1990
|
|
1FQ4
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN HYDROXYETHYLENE INHIBITOR CP-108,420 AND YEAST ASPARTIC PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(2R)-1-{[(2S,3R,5R)-1-cyclohexyl-3-hydroxy-5-{[2-(morpholin-4-yl)ethyl]carbamoyl}oct-7-yn-2-yl]amino}-3-(methylsulfa nyl)-1-oxopropan-2-yl]-1H-benzimidazole-2-carboxamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
1FQ5
 
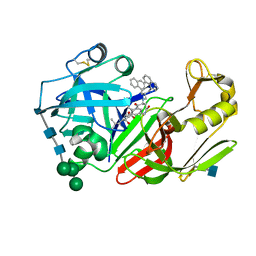 | | X-ray structure of a cyclic statine inhibitor PD-129,541 bound to yeast proteinase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(5S,9S,10S,13S)-9-hydroxy-5,10-bis(2-methylpropyl)-4,7,12,16-tetraoxo-3,6,11,17-tetraazabicyclo[17.3.1]tricosa-1(23),19,21-trien-13-yl]-3-(naphthalen-1-yl)-2-(naphthalen-1-ylmethyl)propanamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
5TOH
 
 | | Crystal Structure of the Marburg Virus VP35 Oligomerization Domain I2 | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Bruhn, J.F, Kirchdoerfer, R.N, Tickle, I.J, Bricogne, G, Saphire, E.O. | | Deposit date: | 2016-10-17 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of the Marburg Virus VP35 Oligomerization Domain.
J. Virol., 91, 2017
|
|
5TOI
 
 | |
1WCC
 
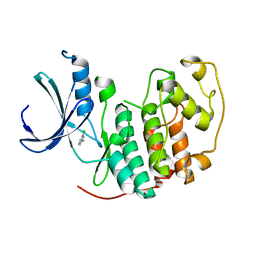 | | Screening for fragment binding by X-ray crystallography | | Descriptor: | 2-AMINO-6-CHLOROPYRAZINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Cleasby, A, O'Reilly, M, Hartshorn, M.J, Murray, C.W, Tickle, I.J, Jhoti, H, Frederickson, M. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
1W0G
 
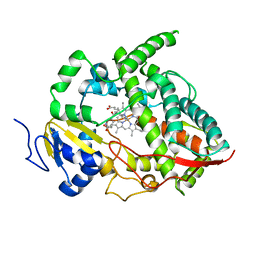 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, METYRAPONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
1W0F
 
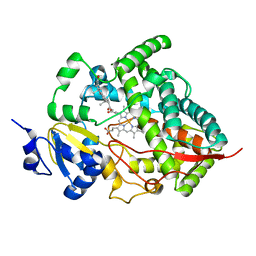 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, PROGESTERONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
1WBU
 
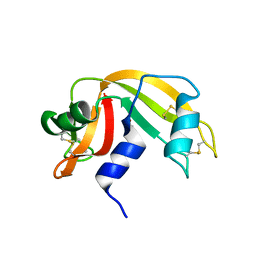 | | Fragment based lead discovery using crystallography | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, RIBONUCLEASE | | Authors: | Cleasby, A, Hartshorn, M.J, Murray, C.W, Jhoti, H, Tickle, I.J. | | Deposit date: | 2004-11-05 | | Release date: | 2005-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
4CMS
 
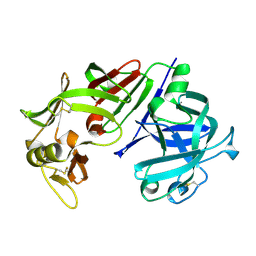 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES IV. STRUCTURE AND REFINEMENT AT 2.2 ANGSTROMS RESOLUTION OF BOVINE CHYMOSIN | | Descriptor: | CHYMOSIN B | | Authors: | Newman, M, Frazao, C, Khan, G, Tickle, I.J, Blundell, T.L, Safro, M, Andreeva, N, Zdanov, A. | | Deposit date: | 1991-11-01 | | Release date: | 1991-11-07 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray analyses of aspartic proteinases. IV. Structure and refinement at 2.2 A resolution of bovine chymosin.
J.Mol.Biol., 221, 1991
|
|
1GN2
 
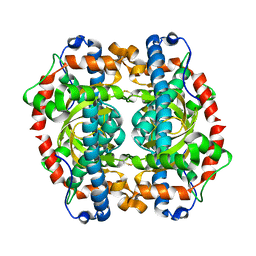 | | S123C mutant of the iron-superoxide dismutase from Mycobacterium tuberculosis. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Tickle, I.J, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Engineering of an Intersubunit Disulfide Bridge in the Iron-Superoxide Dismutase of Mycobacterium Tuberculosis.
Arch.Biochem.Biophys., 397, 2002
|
|
4CXI
 
 | | BTB domain of KEAP1 | | Descriptor: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Cleasby, A, Yon, J, Day, P.J, Richardson, C, Tickle, I.J, Williams, P.A, Callahan, J.F, Carr, R, Concha, N, Kerns, J.K, Qi, H, Sweitzer, T, Ward, P, Davies, T.G. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Btb Domain of Keap1 and its Interaction with the Triterpenoid Antagonist Cddo.
Plos One, 9, 2014
|
|
3CMS
 
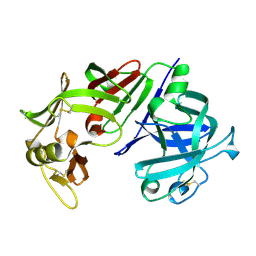 | | ENGINEERING ENZYME SUB-SITE SPECIFICITY: PREPARATION, KINETIC CHARACTERIZATION AND X-RAY ANALYSIS AT 2.0-ANGSTROMS RESOLUTION OF VAL111PHE SITE-MUTATED CALF CHYMOSIN | | Descriptor: | CHYMOSIN B | | Authors: | Newman, M, Frazao, C, Shearer, A, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1990-02-26 | | Release date: | 1992-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering enzyme subsite specificity: preparation, kinetic characterization, and X-ray analysis at 2.0-A resolution of Val111Phe site-mutated calf chymosin.
Biochemistry, 29, 1990
|
|
4CXJ
 
 | | BTB domain of KEAP1 C151W mutant | | Descriptor: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Cleasby, A, Yon, J, Day, P.J, Richardson, C, Tickle, I.J, Williams, P.A, Callahan, J.F, Carr, R, Concha, N, Kerns, J.K, Qi, H, Sweitzer, T, Ward, P, Davies, T.G. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Btb Domain of Keap1 and its Interaction with the Triterpenoid Antagonist Cddo.
Plos One, 9, 2014
|
|
4CXT
 
 | | BTB domain of KEAP1 in complex with CDDO | | Descriptor: | (13alpha,18alpha)-2-cyano-3-hydroxy-12-oxooleana-2,9(11)-dien-28-oic acid, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Cleasby, A, Yon, J, Day, P.J, Richardson, C, Tickle, I.J, Williams, P.A, Callahan, J.F, Carr, R, Concha, N, Kerns, J.K, Qi, H, Sweitzer, T, Ward, P, Davies, T.G. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of the Btb Domain of Keap1 and its Interaction with the Triterpenoid Antagonist Cddo.
Plos One, 9, 2014
|
|
5FPR
 
 | | Structure of Bacterial DNA Ligase with small-molecule ligand pyrimidin-2-amine (AT371) in an alternate binding site. | | Descriptor: | DNA LIGASE, PYRIMIDIN-2-AMINE, SULFATE ION | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPD
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand pyrazine-2-carboxamide (AT513) in an alternate binding site. | | Descriptor: | HEAT SHOCK-RELATED 70KDA PROTEIN 2, PYRAZINE-2-CARBOXAMIDE | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-28 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPM
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 5-phenyl-1,3,4-oxadiazole-2-thiol (AT809) in an alternate binding site. | | Descriptor: | 5-PHENYL-1,3,4-OXADIAZOLE-2-THIOL, HEAT SHOCK-RELATED 70KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPO
 
 | | Structure of Bacterial DNA Ligase with small-molecule ligand 1H- indazol-7-amine (AT4213) in an alternate binding site. | | Descriptor: | 1H-indazol-7-amine, DNA LIGASE | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
