6WF4
 
 | | Crystal Structure of TerC Co-crystallized with Polyporic Acid | | Descriptor: | (2~5~S)-2~3~,2~5~,2~6~-trihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~(2~5~H)-one, ISOPROPYL ALCOHOL, Terfestatin Biosyntheis Enzyme C | | Authors: | Clinger, J.A, Miller, M.D, Hall, R.E, Zhang, Y, Elshahawi, S.I, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional characterization of two cooperative enzymes responsible for the stability
of p-terphenyls.
To be published
|
|
1ZXF
 
 | | Solution structure of a self-sacrificing resistance protein, CalC from Micromonospora echinospora | | Descriptor: | CalC | | Authors: | Singh, S, Hager, M.H, Zhang, C, Griffith, B.R, Lee, M.S, Hallenga, K, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
4WS9
 
 | | Crystal structure of sMAT N159G from Sulfolobus solfataricus | | Descriptor: | PHOSPHATE ION, S-adenosylmethionine synthase | | Authors: | Wang, F, Brady, E.L, Singh, S, Clinger, J.A, Huber, T.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of sMAT N159G from Sulfolobus solfataricus.
To Be Published
|
|
3BUJ
 
 | | Crystal Structure of CalO2 | | Descriptor: | CalO2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McCoy, J.G, Johnson, H.D, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural characterization of CalO2: a putative orsellinic acid P450 oxidase in the calicheamicin biosynthetic pathway.
Proteins, 74, 2009
|
|
3BUS
 
 | | Crystal Structure of RebM | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCoy, J.G, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-03 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of the rebeccamycin sugar 4'-O-methyltransferase RebM.
J.Biol.Chem., 283, 2008
|
|
6UK5
 
 | | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis | | Descriptor: | ACETATE ION, CalS10, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Alvarado, S.K, Miller, M.D, Xu, W, Wang, Z, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis
To Be Published
|
|
4XQ2
 
 | | Ensemble refinement of cystathione gamma lyase (CalE6) D7G from Micromonospora echinospora | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, CalE6, ... | | Authors: | Wang, F, Yennamalli, R.M, Singh, S, Tan, K, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora
To Be Published
|
|
4XR9
 
 | | Crystal structure of CalS8 from Micromonospora echinospora cocrystallized with NAD and TDP-glucose | | Descriptor: | CalS8, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CalS8 from Micromonospora echinospora
To Be Published
|
|
6P9V
 
 | | Crystal Structure of hMAT Mutant K289L | | Descriptor: | ADENOSINE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Miller, M.D, Xu, W, Huber, T.D, Clinger, J.A, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Methionine Adenosyltransferase Engineering to Enable Bioorthogonal Platforms for AdoMet-Utilizing Enzymes.
Acs Chem.Biol., 15, 2020
|
|
4XAU
 
 | | Crystal structure of AtS13 from Actinomadura melliaura | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative aminotransferase | | Authors: | Wang, F, Singh, S, Xu, W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0012 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
4XRR
 
 | | Crystal structure of cals8 from micromonospora echinospora (P294S mutant) | | Descriptor: | CalS8, GLYCEROL | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Characterization of CalS8, a TDP-alpha-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis.
J. Biol. Chem., 290, 2015
|
|
7MSY
 
 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CALCIUM ION, CHLORIDE ION, CalU17, ... | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7ML6
 
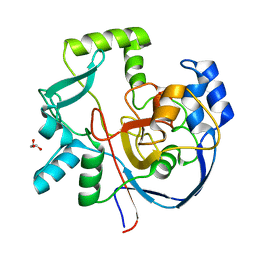 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, GLYCEROL | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
5INJ
 
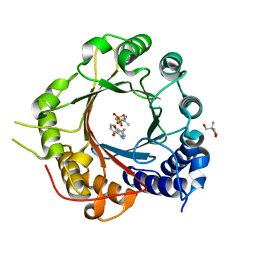 | | Crystal Structure of Prenyltransferase PriB Ternary Complex with L-Tryptophan and Dimethylallyl thiolodiphosphate (DMSPP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Prenyltransferase, ... | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-03-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5HOQ
 
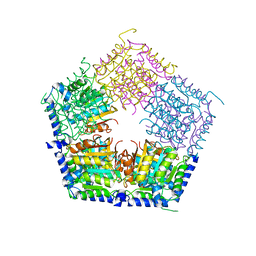 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
2GKD
 
 | |
6ND7
 
 | | The crystal structure of TerB co-crystallized with polyporic acid | | Descriptor: | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6N04
 
 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | Descriptor: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-11-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
5JR3
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Johnson, B.R, Huber, T.D, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-05 | | Release date: | 2016-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone (to be published)
To Be Published
|
|
5K9M
 
 | | Crystal Structure of PriB Binary Complex with Product Diphosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYROPHOSPHATE 2-, PriB Prenyltransferase | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-06-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5JXM
 
 | | Crystal Structure of Prenyltransferase PriB Apo Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PriB | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
4ZAH
 
 | | Crystal structure of sugar aminotransferase WecE with External Aldimine VII from Escherichia coli K-12 | | Descriptor: | [[(2R,3S,5R)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R,4S,5R,6R)-6-methyl-5-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3,4-bis(oxidanyl)oxan-2-yl] hydrogen phosphate, dTDP-4-amino-4,6-dideoxygalactose transaminase | | Authors: | Wang, F, Singh, S, Cao, H, Xu, W, Miller, M.D, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Basis for the Stereochemical Control of Amine Installation in Nucleotide Sugar Aminotransferases.
Acs Chem.Biol., 10, 2015
|
|
4ZAS
 
 | | Crystal structure of sugar aminotransferase CalS13 from Micromonospora echinospora | | Descriptor: | CalS13, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, F, Singh, S, Miller, M.D, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure characterization of sugar aminotransferases CalS13 and WecE provides the basis for a unifying structural model for stereochemical outcome.
To Be Published
|
|
6D2V
 
 | | Apo Structure of TerB, an NADP Dependent Oxidoreductase in the Terfestatin Biosynthesis Pathway | | Descriptor: | CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOCYANATE ION, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
