1DEL
 
 | |
1DEK
 
 | |
4YDY
 
 | |
2PRD
 
 | |
4YDW
 
 | |
1THM
 
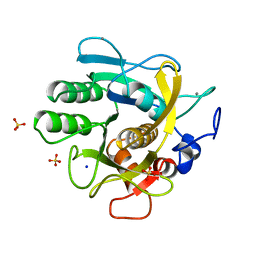 | | CRYSTAL STRUCTURE OF THERMITASE AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, SODIUM ION, SULFATE ION, ... | | Authors: | Teplyakov, A.V, Kuranova, I.P, Harutyunyan, E.H. | | Deposit date: | 1992-02-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of thermitase at 1.4 A resolution.
J.Mol.Biol., 214, 1990
|
|
5KNG
 
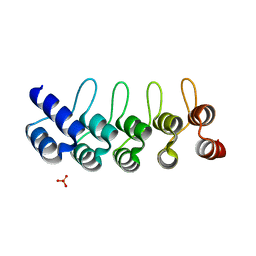 | | CRYSTAL STRUCTURE OF ANTI-IL-13 DARPIN 6G9 | | Descriptor: | DARPIN 6G9, GLYCEROL, PHOSPHATE ION | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Gilliland, G. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational flexibility of an anti-IL-13 DARPin.
Protein Eng. Des. Sel., 30, 2017
|
|
5KNH
 
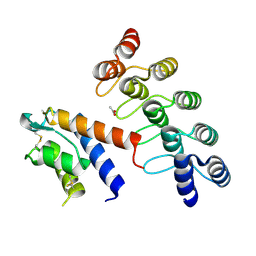 | | CRYSTAL STRUCTURE OF DARPIN 6G9 IN COMPLEX WITH CYNO IL-13 | | Descriptor: | ACETATE ION, DARPIN 6G9, IL13 | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Gilliland, G. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational flexibility of an anti-IL-13 DARPin.
Protein Eng. Des. Sel., 30, 2017
|
|
6AL5
 
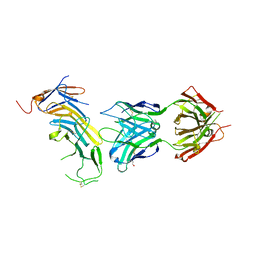 | | COMPLEX BETWEEN CD19 (N138Q MUTANT) AND B43 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-lymphocyte antigen CD19, B43 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of B-cell co-receptor CD19 in complex with antibody B43 reveals an unexpected fold.
Proteins, 86, 2018
|
|
6AL4
 
 | |
4HAG
 
 | | Crystal structure of fc-fragment of human IgG2 antibody (centered crystal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-2 chain C region | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Zhao, Y, Gilliland, G. | | Deposit date: | 2012-09-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | IgG2 Fc structure and the dynamic features of the IgG CH2-CH3 interface.
Mol.Immunol., 56, 2013
|
|
2CTB
 
 | | THE HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CARBOXYPEPTIDASE A AND L-PHENYL LACTATE | | Descriptor: | CARBOXYPEPTIDASE A, ZINC ION | | Authors: | Teplyakov, A, Wilson, K.S, Orioli, P, Mangani, S. | | Deposit date: | 1993-04-02 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structure of the complex between carboxypeptidase A and L-phenyl lactate.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
2CTC
 
 | | THE HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CARBOXYPEPTIDASE A AND L-PHENYL LACTATE | | Descriptor: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, CARBOXYPEPTIDASE A, ZINC ION | | Authors: | Teplyakov, A, Wilson, K.S, Orioli, P, Mangani, S. | | Deposit date: | 1993-04-02 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure of the complex between carboxypeptidase A and L-phenyl lactate.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1PB0
 
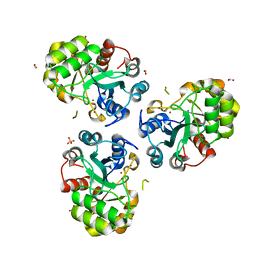 | | YCDX PROTEIN IN AUTOINHIBITED STATE | | Descriptor: | FORMIC ACID, Hypothetical protein ycdX, SULFATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Autoregulation of YcdX protein
To be Published
|
|
4PS4
 
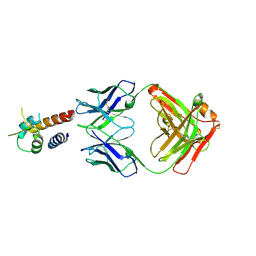 | | Crystal structure of the complex between IL-13 and M1295 FAB | | Descriptor: | Interleukin-13, M1295 HEAVY CHAIN, M1295 LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2014-03-06 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Framework Adaptation of a Mouse Anti-Human Il-13 Antibody.
J.Mol.Biol., 398, 2010
|
|
1MOR
 
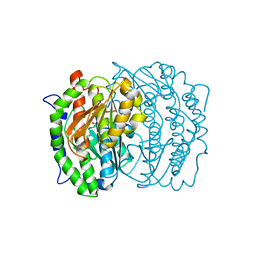 | |
1MOQ
 
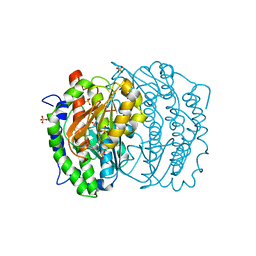 | | ISOMERASE DOMAIN OF GLUCOSAMINE 6-PHOSPHATE SYNTHASE COMPLEXED WITH GLUCOSAMINE 6-PHOSPHATE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, ... | | Authors: | Teplyakov, A. | | Deposit date: | 1997-04-11 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Involvement of the C terminus in intramolecular nitrogen channeling in glucosamine 6-phosphate synthase: evidence from a 1.6 A crystal structure of the isomerase domain.
Structure, 6, 1998
|
|
1MOS
 
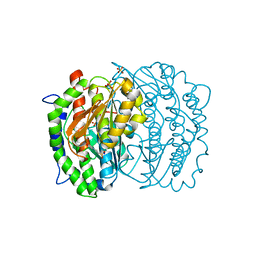 | | ISOMERASE DOMAIN OF GLUCOSAMINE 6-PHOSPHATE SYNTHASE COMPLEXED WITH 2-AMINO-2-DEOXYGLUCITOL 6-PHOSPHATE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE SYNTHASE, ... | | Authors: | Teplyakov, A, Obmolova, G, Badet-Denisot, M.A, Badet, B. | | Deposit date: | 1998-07-15 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of sugar phosphate isomerization by glucosamine 6-phosphate synthase.
Protein Sci., 8, 1999
|
|
3G6A
 
 | |
3G6D
 
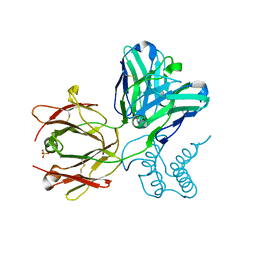 | | Crystal structure of the complex between CNTO607 Fab and IL-13 | | Descriptor: | CNTO607 Fab Heavy chain, CNTO607 Fab Light chain, Interleukin-13, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2009-02-06 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Epitope mapping of anti-interleukin-13 neutralizing antibody CNTO607.
J.Mol.Biol., 389, 2009
|
|
3I2C
 
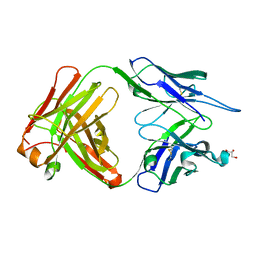 | | Crystal structure of anti-IL-23 antibody CNTO4088 | | Descriptor: | CNTO4088 HEAVY CHAIN, CNTO4088 LIGHT CHAIN, GLYCEROL | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Noncanonical conformation of CDR L1 in the anti-IL-23 antibody CNTO4088.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1S4C
 
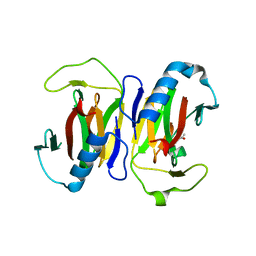 | | YHCH PROTEIN (HI0227) COPPER COMPLEX | | Descriptor: | ACETATE ION, COPPER (II) ION, Protein HI0227 | | Authors: | Teplyakov, A, Obmolova, G, Toedt, J, Gilliland, G.L. | | Deposit date: | 2004-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the bacterial YhcH protein indicates a role in sialic acid catabolism.
J.Bacteriol., 187, 2005
|
|
1RW1
 
 | | YFFB (PA3664) PROTEIN | | Descriptor: | ISOPROPYL ALCOHOL, conserved hypothetical protein yffB | | Authors: | Teplyakov, A, Pullalarevu, S, Obmolova, G, Doseeva, V, Galkin, A, Herzberg, O, Dauter, M, Dauter, Z, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-15 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
Bmc Struct.Biol., 4, 2004
|
|
1ZLP
 
 | | Petal death protein PSR132 with cysteine-linked glutaraldehyde forming a thiohemiacetal adduct | | Descriptor: | 5-HYDROXYPENTANAL, MAGNESIUM ION, petal death protein | | Authors: | Teplyakov, A, Liu, S, Lu, Z, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2005-05-08 | | Release date: | 2006-01-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Petal Death Protein from Carnation Flower.
Biochemistry, 44, 2005
|
|
1R45
 
 | | ADP-ribosyltransferase C3bot2 from Clostridium botulinum, triclinic form | | Descriptor: | GLYCEROL, Mono-ADP-ribosyltransferase C3, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L, Narumiya, S. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of ADP-ribosyltransferase C3bot2 from Clostridium botulinum
To be Published
|
|
