1AU9
 
 | | SUBTILISIN BPN' MUTANT 8324 IN CITRATE | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SUBTILISIN BPN', ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-09-12 | | Release date: | 1997-12-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1AOE
 
 | | CANDIDA ALBICANS DIHYDROFOLATE REDUCTASE COMPLEXED WITH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE (NADPH) AND 1,3-DIAMINO-7-(1-ETHYEPROPYE)-7H-PYRRALO-[3,2-F]QUINAZOLINE (GW345) | | Descriptor: | 7-(1-ETHYL-PROPYL)-7H-PYRROLO-[3,2-F]QUINAZOLINE-1,3-DIAMINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Whitlow, M, Howard, A.J, Stewart, D. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-07 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of Candida albicans dihydrofolate reductase. High resolution structures of the holoenzyme and an inhibited ternary complex.
J.Biol.Chem., 272, 1997
|
|
1AI9
 
 | | CANDIDA ALBICANS DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Whitlow, M, Howard, A.J, Stewart, D. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystallographic studies of Candida albicans dihydrofolate reductase. High resolution structures of the holoenzyme and an inhibited ternary complex.
J.Biol.Chem., 272, 1997
|
|
1AK9
 
 | | SUBTILISIN MUTANT 8321 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-05-30 | | Release date: | 1997-11-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1A2Q
 
 | | SUBTILISIN BPN' MUTANT 7186 | | Descriptor: | ACETONE, CALCIUM ION, SUBTILISIN BPN' | | Authors: | Gilliland, G.L, Whitlow, M, Howard, A.J. | | Deposit date: | 1998-01-08 | | Release date: | 1998-04-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1AQN
 
 | | SUBTILISIN MUTANT 8324 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SUBTILISIN 8324, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-07-31 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stabilityfor subtilisin BPN' through incremental changes in the free energy of unfolding
To be published
|
|
3TM9
 
 | | Y29A mutant of Vitreoscilla stercoraria hemoglobin | | Descriptor: | 1,2-ETHANEDIOL, Bacterial hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ratakonda, S, Anand, A, Dikshit, K, Stark, B.C, Howard, A.J. | | Deposit date: | 2011-08-31 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystallographic structure determination of B10 mutants of Vitreoscilla hemoglobin: role of Tyr29 (B10) in the structure of the ligand-binding site.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3XIS
 
 | |
3TM3
 
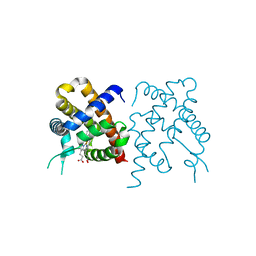 | | Wild-type hemoglobin from Vitreoscilla stercoraria | | Descriptor: | Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ratakonda, S, Anand, A, Dikshit, K, Stark, B.C, Howard, A.J. | | Deposit date: | 2011-08-31 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic structure determination of B10 mutants of Vitreoscilla hemoglobin: role of Tyr29 (B10) in the structure of the ligand-binding site.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3TLD
 
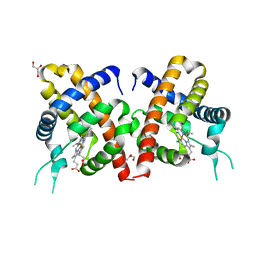 | | Crystal Structure of Y29F mutant of Vitreoscilla hemoglobin | | Descriptor: | Bacterial hemoglobin, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ratakonda, S, Anand, A, Dikshit, K, Stark, B.C, Howard, A.J. | | Deposit date: | 2011-08-29 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystallographic structure determination of B10 mutants of Vitreoscilla hemoglobin: role of Tyr29 (B10) in the structure of the ligand-binding site.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2RE9
 
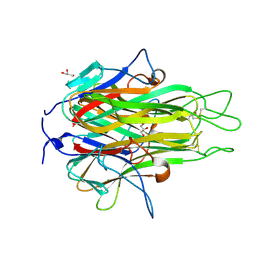 | | Crystal structure of TL1A at 2.1 A | | Descriptor: | GLYCEROL, MAGNESIUM ION, TNF superfamily ligand TL1A | | Authors: | Jin, T.C, Guo, F, Kim, S, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of TNF ligand family member TL1A at 2.1 A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
2XIS
 
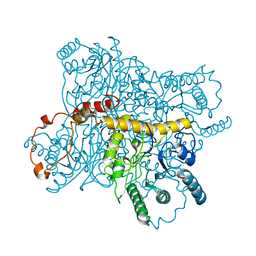 | |
2NML
 
 | | Crystal structure of HEF2/ERH at 1.55 A resolution | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Jin, T.C, Guo, F, Serebriiskii, I.G, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2006-10-21 | | Release date: | 2006-10-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55 A resolution X-ray crystal structure of HEF2/ERH and insights into its transcriptional and cell-cycle interaction networks.
Proteins, 68, 2007
|
|
4QMJ
 
 | | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array | | Descriptor: | Cytoskeleton-associated protein 5 | | Authors: | Fox, J.C, Howard, A.E, Currie, J.D, Rogers, S.L, Slep, K.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array.
Mol.Biol.Cell, 25, 2014
|
|
4QMI
 
 | | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array | | Descriptor: | Cytoskeleton-associated protein 5 | | Authors: | Fox, J.C, Howard, A.E, Currie, J.D, Rogers, S.L, Slep, K.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array.
Mol.Biol.Cell, 25, 2014
|
|
4QMH
 
 | | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array | | Descriptor: | LP04448p, SULFATE ION | | Authors: | Fox, J.C, Howard, A.E, Currie, J.D, Rogers, S.L, Slep, K.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array.
Mol.Biol.Cell, 25, 2014
|
|
2O0O
 
 | | Crystal structure of TL1A | | Descriptor: | MAGNESIUM ION, TNF superfamily ligand TL1A | | Authors: | Jin, T.C, Kim, S, Guo, F, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2006-11-27 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of TNF ligand family member TL1A at 2.1A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
1G1B
 
 | | CHORISMATE LYASE (WILD-TYPE) WITH BOUND PRODUCT | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Kim, K.J, Howard, A, Vilker, V.L. | | Deposit date: | 2000-10-11 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
1XIS
 
 | |
1TT8
 
 | | CHORISMATE LYASE WITH PRODUCT, 1.0 A RESOLUTION | | Descriptor: | Chorismate-pyruvate lyase, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Vilker, V, Howard, A. | | Deposit date: | 2004-06-22 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural analysis of ligand binding and catalysis in chorismate lyase.
Arch.Biochem.Biophys., 445, 2006
|
|
4XIS
 
 | |
1X6I
 
 | | Crystal structure of ygfY from Escherichia coli | | Descriptor: | Hypothetical protein ygfY | | Authors: | Lim, K, Doseeva, V, Sarikaya Demirkan, E, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation
Proteins, 58, 2005
|
|
1ZLP
 
 | | Petal death protein PSR132 with cysteine-linked glutaraldehyde forming a thiohemiacetal adduct | | Descriptor: | 5-HYDROXYPENTANAL, MAGNESIUM ION, petal death protein | | Authors: | Teplyakov, A, Liu, S, Lu, Z, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2005-05-08 | | Release date: | 2006-01-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Petal Death Protein from Carnation Flower.
Biochemistry, 44, 2005
|
|
1X6J
 
 | | Crystal structure of ygfY from Escherichia coli | | Descriptor: | Hypothetical protein ygfY | | Authors: | Lim, K, Doseeva, V, Sarikaya Demirkan, E, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation
Proteins, 58, 2005
|
|
4Y5J
 
 | | Drosophila melanogaster Mini spindles TOG3 | | Descriptor: | MINI SPINDLES TOG3 | | Authors: | Slep, K.C, Howard, A.E. | | Deposit date: | 2015-02-11 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Drosophila melanogaster Mini Spindles TOG3 Utilizes Unique Structural Elements to Promote Domain Stability and Maintain a TOG1- and TOG2-like Tubulin-binding Surface.
J.Biol.Chem., 290, 2015
|
|
