6IR6
 
 | | Green fluorescent protein variant GFPuv with the native lysine residue at the C-terminus | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Nakatani, T, Yasui, N, Yamashita, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
6IR7
 
 | | Green fluorescent protein variant GFPuv with the modification to 6-hydroxynorleucine at the C-terminus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-HYDROXY-L-NORLEUCINE, Green fluorescent protein, ... | | Authors: | Nakatani, T, Yasui, N, Yamashita, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.277 Å) | | Cite: | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
2D2I
 
 | | Crystal Structure of NADP-Dependent Glyceraldehyde-3-Phosphate Dehydrogenase from Synechococcus Sp. complexed with Nadp+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, glyceraldehyde 3-phosphate dehydrogenase | | Authors: | Kitatani, T, Nakamura, Y, Wada, K, Kinoshita, T, Tamoi, M, Shigeoka, S, Tada, T. | | Deposit date: | 2005-09-09 | | Release date: | 2006-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Synechococcus PCC7942 complexed with NADP
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2ZWI
 
 | | Crystal structure of alpha/beta-Galactoside alpha-2,3-Sialyltransferase from a Luminous Marine Bacterium, Photobacterium phosphoreum | | Descriptor: | Alpha-/beta-galactoside alpha-2,3-sialyltransferase, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Iwatani, T, Okino, N, Sakakura, M, Kajiwara, H, Ichikawa, M, Takakura, Y, Kimura, M, Ito, M, Yamamoto, T, Kakuta, Y. | | Deposit date: | 2008-12-05 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of alpha/beta-galactoside alpha2,3-sialyltransferase from a luminous marine bacterium, Photobacterium phosphoreum
Febs Lett., 583, 2009
|
|
2DUU
 
 | | Crystal Structure of apo-form of NADP-Dependent Glyceraldehyde-3-Phosphate Dehydrogenase from Synechococcus Sp. | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, SULFATE ION | | Authors: | Kitatani, T, Nakamura, Y, Wada, K, Kinoshita, T, Tamoi, M, Shigeoka, S, Tada, T. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of apo-glyceraldehyde-3-phosphate dehydrogenase from Synechococcus PCC7942
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2CYM
 
 | | EFFECTS OF AMINO ACID SUBSTITUTION ON THREE-DIMENSIONAL STRUCTURE: AN X-RAY ANALYSIS OF CYTOCHROME C3 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH AT 2 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Morimoto, Y, Tani, T, Okumura, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1993-09-29 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of amino acid substitution on three-dimensional structure: an X-ray analysis of cytochrome c3 from Desulfovibrio vulgaris Hildenborough at 2 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
1VFP
 
 | |
2QUL
 
 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii at 1.79 A resolution | | Descriptor: | D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
2QUM
 
 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii with D-tagatose | | Descriptor: | D-tagatose, D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
6LK4
 
 | | Crystal structure of GMP reductase from Trypanosoma brucei in complex with guanosine 5'-triphosphate | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Guanosine 5'-monophosphate Reductase, PHOSPHATE ION | | Authors: | Mase, H, Otani, T, Imamura, A, Nishimura, S, Inui, T. | | Deposit date: | 2019-12-18 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-beta-synthase domain.
Nat Commun, 11, 2020
|
|
2IRF
 
 | | CRYSTAL STRUCTURE OF AN IRF-2/DNA COMPLEX. | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*TP*TP*TP*CP*AP*CP*(5IU)P*T)-3'), DNA (5'-D(P*AP*AP*GP*TP*GP*AP*AP*AP*GP*(5IU)P*GP*A)-3'), INTERFERON REGULATORY FACTOR 2, ... | | Authors: | Fujii, Y, Shimizu, T, Kusumoto, M, Kyogoku, Y, Taniguchi, T, Hakoshima, T. | | Deposit date: | 1999-05-30 | | Release date: | 1999-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences.
EMBO J., 18, 1999
|
|
5AX6
 
 | | The crystal structure of CofB, the minor pilin subunit of CFA/III from human enterotoxigenic Escherichia coli. | | Descriptor: | ACETATE ION, CofB | | Authors: | Kawahara, K, Oki, K, Fukaksua, F, Maruno, T, Kobayashi, Y, Daisuke, M, Taniguchi, T, Honda, T, Iida, T, Nakamura, S, Ohkubo, T. | | Deposit date: | 2015-07-16 | | Release date: | 2016-03-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Homo-trimeric Structure of the Type IVb Minor Pilin CofB Suggests Mechanism of CFA/III Pilus Assembly in Human Enterotoxigenic Escherichia coli
J.Mol.Biol., 428, 2016
|
|
3W81
 
 | | Human alpha-l-iduronidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Maita, N, Tsukimura, T, Taniguchi, T, Saito, S, Ohno, K, Taniguchi, H, Sakuraba, H. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human alpha-L-iduronidase uses its own N-glycan as a substrate-binding and catalytic module
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7FGK
 
 | | The Fab antibody single structure against tau protein. | | Descriptor: | Fab Heavy Chain, Fab Light Chain, GLYCEROL, ... | | Authors: | Tsuchida, T, Susa, K, Kibiki, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The free structure of the Fab domain of antibody that recognizes the PHF core region VQIINK in Tau protein.
To Be Published
|
|
7FGJ
 
 | | The cross-reaction complex structure with VQILNK peptide and the tau antibody's Fab domain. | | Descriptor: | CHLORIDE ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The cross-reaction complex structure with VQILNK peptide and the antibody's Fab domain.
To Be Published
|
|
7FGL
 
 | | The complex structure of PHF core domain peptide of tau, VQIVYK, and antibody's Fab domain. | | Descriptor: | AMMONIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cross-reaction complex structure with VQIVYK of tau and the antibody's Fab domain.
To Be Published
|
|
3W82
 
 | | Human alpha-L-iduronidase in complex with iduronic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Maita, N, Tsukimura, T, Taniguchi, T, Saito, S, Ohno, K, Taniguchi, H, Sakuraba, H. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Human alpha-L-iduronidase uses its own N-glycan as a substrate-binding and catalytic module
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7FGR
 
 | | The cross-reaction complex structure with VQIFNK peptide and the tau antibody's Fab domain. | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Fab Heavy Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The cross-reaction complex structure with VQIFNK peptide and the tau antibody's Fab domain.
To Be Published
|
|
1VBL
 
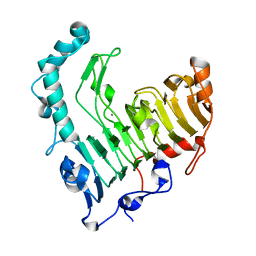 | | Structure of the thermostable pectate lyase PL 47 | | Descriptor: | CALCIUM ION, pectate lyase 47 | | Authors: | Nakaniwa, T, Tada, T, Yamaguchi, A, Kitatani, T, Takao, M, Sakai, T, Nishimura, K. | | Deposit date: | 2004-02-27 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Thermostability of Pectate Lyase from Bacillus sp. TS 47
To be Published
|
|
1WL7
 
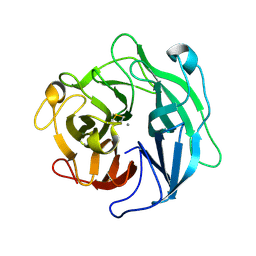 | | Structure of the thermostable arabinanase | | Descriptor: | CALCIUM ION, arabinanase-TS | | Authors: | Yamaguchi, A, Tada, T, Nakaniwa, T, Kitatani, T. | | Deposit date: | 2004-06-21 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for thermostability of endo-1,5-alpha-L-arabinanase from Bacillus thermodenitrificans TS-3.
J.Biochem.(Tokyo), 137, 2005
|
|
5B22
 
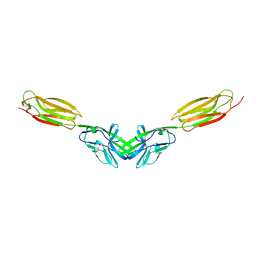 | | Dimer structure of murine Nectin-3 D1D2 | | Descriptor: | Nectin-3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Takebe, K, Sangawa, T, Katsutani, T, Narita, H, Suzuki, M. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Dimer structure of murine Nectin-3 D1D2
To Be Published
|
|
3VOR
 
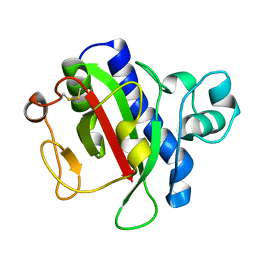 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1HZK
 
 | |
2QUN
 
 | | Crystal Structure of D-tagatose 3-epimerase from Pseudomonas cichorii in Complex with D-fructose | | Descriptor: | D-fructose, D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
1IRG
 
 | | INTERFERON REGULATORY FACTOR-2 DNA BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | INTERFERON REGULATORY FACTOR-2 | | Authors: | Furui, J, Uegaki, K, Yamazaki, T, Shirakawa, M, Swindells, M.B, Harada, H, Taniguchi, T, Kyogoku, Y. | | Deposit date: | 1997-11-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IRF-2 DNA-binding domain: a novel subgroup of the winged helix-turn-helix family.
Structure, 6, 1998
|
|
