8HOC
 
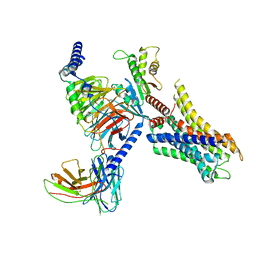 | | Cryo-EM structure of ligand histamine-bound Histamine H4 receptor Gi complex | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tang, W.Q, Sun, X.Y, Li, F.H, Wang, J.Y. | | Deposit date: | 2022-12-09 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure insights into Histamine H4 receptor activation by an endogenous ligand histamine and agonist imetit
To Be Published
|
|
8HN8
 
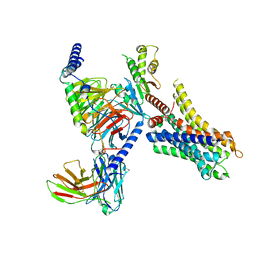 | | Cryo-EM structure of ligand histamine-bound Histamine H4 receptor Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, W.Q, Sun, X.Y, Li, F.H, Wang, J.Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure insights into Histamine H4 receptor activation by an endogenous ligand histamine and agonist imetit
To Be Published
|
|
6VHJ
 
 | |
6XLY
 
 | |
6XOS
 
 | |
6XOU
 
 | |
6XOV
 
 | |
6XOT
 
 | |
6VGT
 
 | |
6VE9
 
 | |
6XOW
 
 | |
8D56
 
 | | One RBD-up state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D5A
 
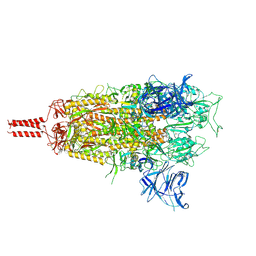 | | Middle state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D55
 
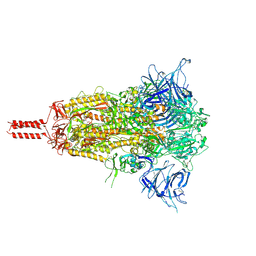 | | Closed state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1F03
 
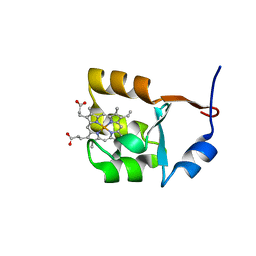 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1PK0
 
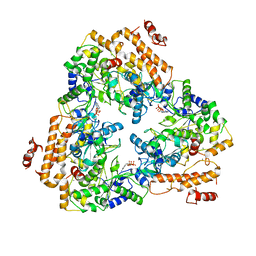 | | Crystal Structure of the EF3-CaM complexed with PMEApp | | Descriptor: | (ADENIN-9-YL-ETHOXYMETHYL)-HYDROXYPHOSPHINYL-DIPHOSPHATE, CALCIUM ION, Calmodulin, ... | | Authors: | Shen, Y, Tang, W.J. | | Deposit date: | 2003-06-04 | | Release date: | 2004-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Selective inhibition of anthrax edema factor by adefovir, a drug for chronic hepatitis B virus infection.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2G54
 
 | |
2G47
 
 | |
2G48
 
 | |
2G49
 
 | |
2G56
 
 | |
2WK3
 
 | |
5L2U
 
 | |
2YPU
 
 | | human insulin degrading enzyme E111Q in complex with inhibitor compound 41367 | | Descriptor: | 2-[[2-[[(2S)-3-(3H-IMIDAZOL-4-YL)-1-METHOXY-1-OXO-PROPAN-2-YL]AMINO]-2-OXO-ETHYL]-(PHENYLMETHYL)AMINO]ETHANOIC ACID, INSULIN-DEGRADING ENZYME, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.-J. | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Imidazole-Derived 2-[N-Carbamoylmethyl-Alkylamino]Acetic Acids,Substrate-Dependent Modulators of Insulin-Degrading Enzyme in Amyloid-Beta Hydrolysis
Eur J Med Chem, 79C, 2014
|
|
