5Z0C
 
 | | Nerol dehydrogenase from Persicaria minor | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Nerol dehydrogenase, ... | | Authors: | Tan, C.S, Ng, C.L, Zainal, Z. | | Deposit date: | 2017-12-19 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural and kinetic studies of a novel nerol dehydrogenase from Persicaria minor, a nerol-specific enzyme for citral biosynthesis.
Plant Physiol. Biochem., 123, 2017
|
|
7RTY
 
 | |
8CZK
 
 | |
8CZL
 
 | | Human LanCL1 bound to methyl glutathione (MeGSH) | | Descriptor: | Glutathione S-transferase LANCL1, L-GAMMA-GLUTAMYL-S-METHYLCYSTEINYLGLYCINE, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D19
 
 | | Human LanCL1 bound to GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D0V
 
 | | Human LanCL1 C264A mutant bound to GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7SUN
 
 | |
2MZY
 
 | | 1H, 13C, and 15N Chemical Shift Assignments and structure of Probable Fe(2+)-trafficking protein from Burkholderia pseudomallei 1710b. | | Descriptor: | Probable Fe(2+)-trafficking protein | | Authors: | Tang, C, Yang, F, Barnwal, R, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Probable Fe(2+)-trafficking protein
To be Published
|
|
2MRC
 
 | |
6JDS
 
 | |
6JDU
 
 | | Crystal structure of PRRSV nsp10 (helicase) | | Descriptor: | CALCIUM ION, PP1b, ZINC ION | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2019-02-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure.
Viruses, 12, 2020
|
|
3J48
 
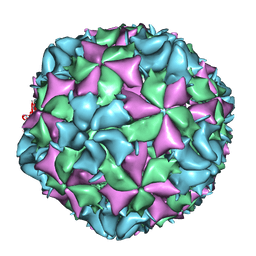 | | Cryo-EM structure of Poliovirus 135S particles | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Butan, C, Fiman, D.J, Hogle, J.M. | | Deposit date: | 2013-06-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-Electron Microscopy Reconstruction Shows Poliovirus 135S Particles Poised for Membrane Interaction and RNA Release.
J.Virol., 88, 2014
|
|
5IW3
 
 | | anti-CD20 monoclonal antibody Fc fragment | | Descriptor: | ACETATE ION, Ig gamma-1 chain C region, SULFATE ION, ... | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of anti-CD20 monoclonal antibody Fc fragment at 2.05 Angstroms resolution
To Be Published
|
|
5IW6
 
 | | anti-CD20 monoclonal antibody Fc fragment | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2016-03-22 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of anti-CD20 monoclonal antibody Fc fragment at 2.34 Angstroms resolution
To Be Published
|
|
6UOT
 
 | |
6UOV
 
 | |
5BK9
 
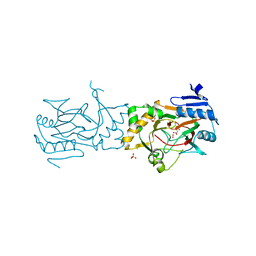 | | AAD-1 Bound to the Vanadyl Ion and Succinate | | Descriptor: | (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, PHOSPHATE ION, SUCCINIC ACID, ... | | Authors: | Ongpipattanakul, C, Chekan, J.R. | | Deposit date: | 2019-06-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7UD6
 
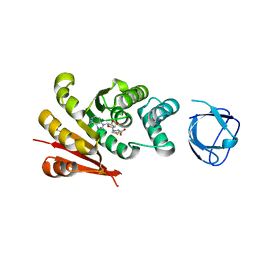 | |
2W9X
 
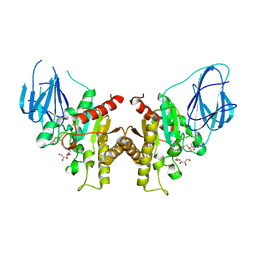 | | The active site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions | | Descriptor: | GLYCEROL, PUTATIVE ACETYL XYLAN ESTERASE | | Authors: | Montanier, C, Money, V.A, Pires, V, Flint, J.E, Benedita, P.A, Goyal, A, Prates, J.A, Izumi, A, Stalbrand, H, Morland, C, Cartmell, A, Kolenova, K, Topakas, E, Dobson, E, Bolam, D.N, Davies, G.J, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
4ZWG
 
 | | Crystal structure of the GTP-dATP-bound catalytic core of SAMHD1 phosphomimetic T592E mutant | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tang, C, Ji, X, Xiong, Y. | | Deposit date: | 2015-05-19 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impaired dNTPase Activity of SAMHD1 by Phosphomimetic Mutation of Thr-592.
J.Biol.Chem., 290, 2015
|
|
4ZWE
 
 | |
1WCR
 
 | |
2XFE
 
 | | vCBM60 in complex with galactobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
1UTY
 
 | |
1L6N
 
 | |
