6EG7
 
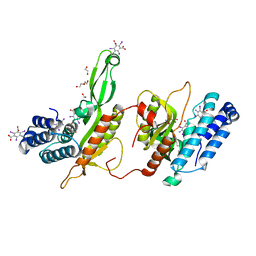 | | BbvCI B2 dimer with I3C clusters | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, BbvCI endonuclease subunit 2, ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2018-08-19 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure, subunit organization and behavior of the asymmetric Type IIT restriction endonuclease BbvCI.
Nucleic Acids Res., 47, 2019
|
|
5JSB
 
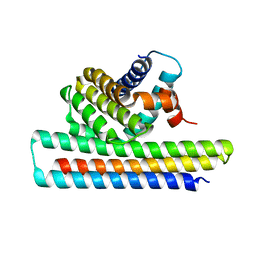 | | Crystal structure of Mcl1-inhibitor complex | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mcl-1 inhibitor | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2016-05-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer.
Elife, 5, 2016
|
|
5JSN
 
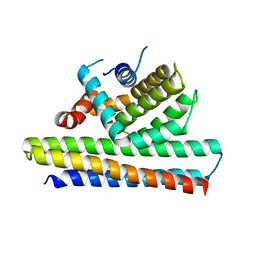 | | Bcl2-inhibitor complex | | Descriptor: | Apoptosis regulator Bcl-2, Bcl2 inhibitor | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2016-05-09 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer.
Elife, 5, 2016
|
|
7RDR
 
 | |
1QVJ
 
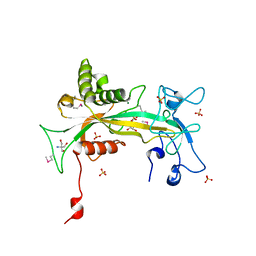 | | structure of NUDT9 complexed with ribose-5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-beta-D-ribofuranose, ADP-ribose pyrophosphatase, ... | | Authors: | Shen, B.W, Perraud, A.-L, Scharenberg, A.S, Stoddard, B.L. | | Deposit date: | 2003-08-27 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The crystal structure and mutational analysis of human NUDT9
J.Mol.Biol., 332, 2003
|
|
1OAT
 
 | | ORNITHINE AMINOTRANSFERASE | | Descriptor: | ORNITHINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shen, B.W, Schirmer, T, Jansonius, J.N. | | Deposit date: | 1997-03-26 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human recombinant ornithine aminotransferase.
J.Mol.Biol., 277, 1998
|
|
5E63
 
 | | K262A mutant of I-SmaMI | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-METHOXYETHANOL, DNA (5'-D(P*CP*AP*GP*GP*TP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Shen, B, Stoddard, B. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
8EMH
 
 | |
8EMC
 
 | |
1U3E
 
 | | DNA binding and cleavage by the HNH homing endonuclease I-HmuI | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 36-MER, ... | | Authors: | Shen, B.W, Landthaler, M, Shub, D.A, Stoddard, B.L. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | DNA Binding and Cleavage by the HNH Homing Endonuclease I-HmuI.
J.Mol.Biol., 342, 2004
|
|
5E5S
 
 | | I-SmaMI K103A mutant | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Bottom strand DNA, DNA (5'-D(P*CP*AP*GP*GP*TP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Shen, B.W. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
5E5O
 
 | |
5E67
 
 | | K103A/K262A double mutant of I-SmaMI | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-METHOXYETHANOL, DNA bottom strand, ... | | Authors: | Shen, B.W, Stoddard, B. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
3LDY
 
 | | An extraordinary mechanism of DNA perturbation exhibited by the rare-cutting HNH restriction endonuclease PacI | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*GP*GP*CP*TP*TP*A)-3'), DNA (5'-D(P*AP*TP*TP*AP*AP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Shen, B.W, Heiter, D, Chan, S.-H, Xu, S.-Y, Wilson, G, Stoddard, B.L. | | Deposit date: | 2010-01-13 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.
Structure, 18, 2010
|
|
3M7K
 
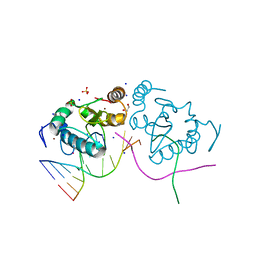 | | Crystal structure of PacI-DNA Enzyme product complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*TP*TP*AP*AP*T)-3'), DNA (5'-D(P*TP*AP*AP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.
Structure, 18, 2010
|
|
5TGX
 
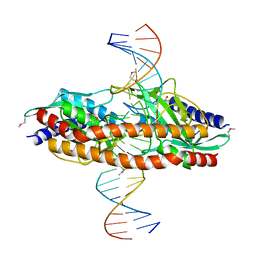 | |
1Q33
 
 | | Crystal structure of human ADP-ribose pyrophosphatase NUDT9 | | Descriptor: | ADP-ribose pyrophosphatase, SULFATE ION, beta-D-glucopyranose | | Authors: | Shen, B.W, Perraud, A.L, Scharenberg, A, Stoddard, B.L. | | Deposit date: | 2003-07-28 | | Release date: | 2003-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Crystal Structure and Mutational Analysis of Human NUDT9
J.Mol.Biol., 332, 2003
|
|
5TH3
 
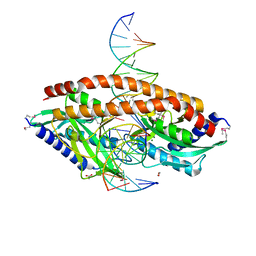 | |
5E5P
 
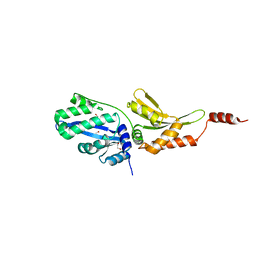 | | Wild type I-SmaMI in the space group of C121 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, I-SmaMI LAGLIDADG meganuclease | | Authors: | Shen, B.W. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
4OYD
 
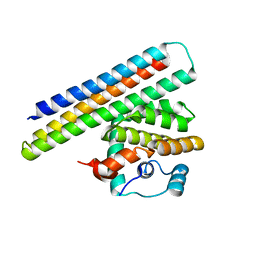 | | Crystal structure of a computationally designed inhibitor of an Epstein-Barr viral Bcl-2 protein | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, Computationally designed Inhibitor | | Authors: | Shen, B, Procko, E, Baker, D, Stoddard, B. | | Deposit date: | 2014-02-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A computationally designed inhibitor of an epstein-barr viral bcl-2 protein induces apoptosis in infected cells.
Cell, 157, 2014
|
|
5TGQ
 
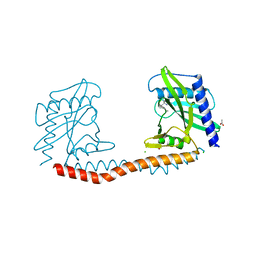 | | Restriction-modification system Type II R.SwaI, DNA free | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Shen, B.W, stoddard, B.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | DNA recognition by the SwaI restriction endonuclease involves unusual distortion of an 8 base pair A:T-rich target.
Nucleic Acids Res., 45, 2017
|
|
7LO5
 
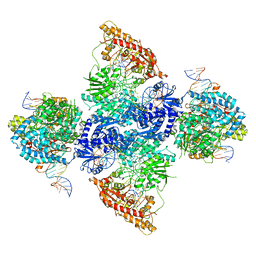 | | cryoEM structure DrdV-DNA complex | | Descriptor: | CALCIUM ION, DNA (27-MER), DNA (28-MER), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2021-02-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Coordination of phage genome degradation versus host genome protection by a bifunctional restriction-modification enzyme visualized by CryoEM.
Structure, 29, 2021
|
|
7LVV
 
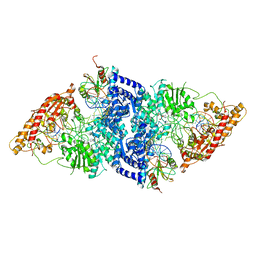 | | cryoEM structure DrdV-DNA complex | | Descriptor: | CALCIUM ION, DNA (27-MER), DNA (28-MER), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Coordination of phage genome degradation versus host genome protection by a bifunctional restriction-modification enzyme visualized by CryoEM.
Structure, 29, 2021
|
|
4QPZ
 
 | | Crystal structure of the formolase FLS_v2 in space group P 21 | | Descriptor: | Formolase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L, Baker, D. | | Deposit date: | 2014-06-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4QQ8
 
 | | Crystal structure of the formolase FLS in space group P 43 21 2 | | Descriptor: | 1,2-ETHANEDIOL, Formolase, MAGNESIUM ION, ... | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L. | | Deposit date: | 2014-06-26 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
