8DVH
 
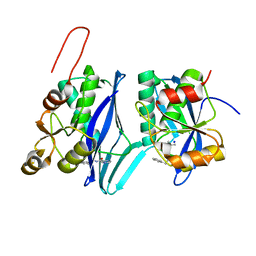 | | Crystal structure of ATP-dependent Lon protease from Bacillus subtillis (BsLonBA) | | Descriptor: | Lon protease 2, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, SODIUM ION | | Authors: | Sekula, B, Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unique Structural Fold of LonBA Protease from Bacillus subtilis, a Member of a Newly Identified Subfamily of Lon Proteases.
Int J Mol Sci, 23, 2022
|
|
4ZBR
 
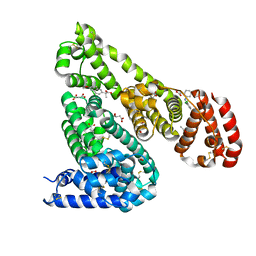 | | Crystal Structure of Equine Serum Albumin in complex with Diclofenac and Naproxen | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, (2S)-2-hydroxybutanedioic acid, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ... | | Authors: | Sekula, B, Bujacz, A, Bujacz, G. | | Deposit date: | 2015-04-15 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Insights into the Competitive Binding of Diclofenac and Naproxen by Equine Serum Albumin.
J.Med.Chem., 59, 2016
|
|
4ZBQ
 
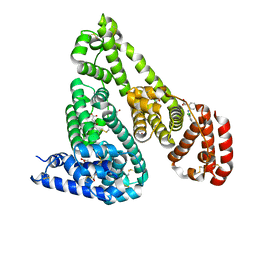 | | Crystal Structure of Equine Serum Albumin in complex with Diclofenac | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ACETATE ION, ... | | Authors: | Sekula, B, Bujacz, A, Bujacz, G. | | Deposit date: | 2015-04-15 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into the Competitive Binding of Diclofenac and Naproxen by Equine Serum Albumin.
J.Med.Chem., 59, 2016
|
|
5DBY
 
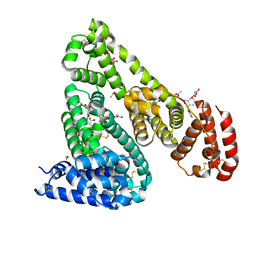 | | Crystal Structure of Equine Serum Albumin in Complex with Diclofenac and Naproxen Obtained in Displacement Experiment | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, (2S)-2-hydroxybutanedioic acid, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ... | | Authors: | Sekula, B, Bujacz, A, Bujacz, G. | | Deposit date: | 2015-08-22 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Insights into the Competitive Binding of Diclofenac and Naproxen by Equine Serum Albumin.
J.Med.Chem., 59, 2016
|
|
4J2V
 
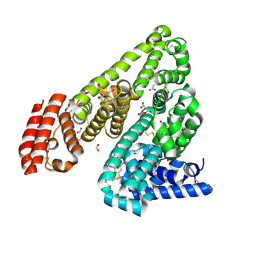 | | Crystal Structure of Equine Serum Albumin in complex with 3,5-diiodosalicylic acid | | Descriptor: | 2-HYDROXY-3,5-DIIODO-BENZOIC ACID, ACETATE ION, FORMIC ACID, ... | | Authors: | Sekula, B, Bujacz, A, Zielinski, K, Bujacz, G. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystallographic studies of the complexes of bovine and equine serum albumin with 3,5-diiodosalicylic acid.
Int.J.Biol.Macromol., 60C, 2013
|
|
6CZY
 
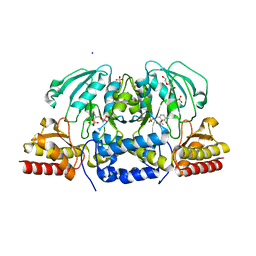 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with Pyridoxamine-5'-phosphate (PMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
6CZX
 
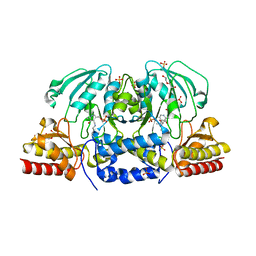 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with PLP internal aldimine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DI(HYDROXYETHYL)ETHER, Phosphoserine aminotransferase 1, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
6CZZ
 
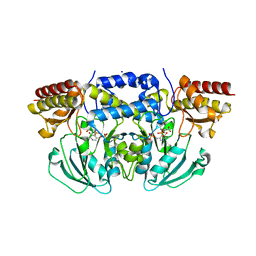 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with PLP-phosphoserine geminal diamine intermediate | | Descriptor: | PHOSPHOSERINE, PYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase 1, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
5ID7
 
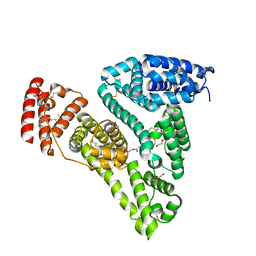 | | Crystal structure of human serum albumin in complex with phosphorodithioate derivative of myristoyl cyclic phosphatidic acid (cPA) | | Descriptor: | (4S)-2-sulfanylidene-4-[(tetradecanoyloxy)methyl]-1,3,2lambda~5~-dioxaphospholane-2-thiolate, DI(HYDROXYETHYL)ETHER, Serum albumin, ... | | Authors: | Sekula, B, Bujacz, A, Rytczak, P, Bujacz, G. | | Deposit date: | 2016-02-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural evidence of the species-dependent albumin binding of the modified cyclic phosphatidic acid with cytotoxic properties.
Biosci.Rep., 36, 2016
|
|
5H8L
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) C158S mutant in complex with putrescine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5H8K
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) C158S mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5H8I
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) in complex with N-(dihydroxymethyl)putrescine | | Descriptor: | (4-azanylbutylamino)methanediol, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
4OT2
 
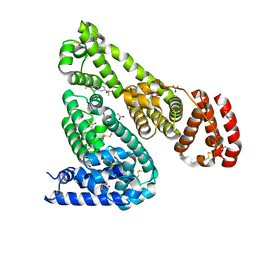 | | Crystal Structure of Equine Serum Albumin in complex with Naproxen | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, ACETATE ION, MALONATE ION, ... | | Authors: | Sekula, B, Bujacz, A, Zielinski, K, Bujacz, G. | | Deposit date: | 2014-02-13 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural studies of bovine, equine, and leporine serum albumin complexes with naproxen.
Proteins, 82, 2014
|
|
5H8J
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) in complex with cadaverine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
6O65
 
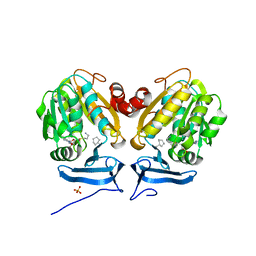 | |
6O63
 
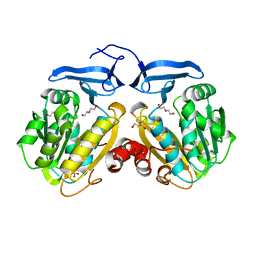 | |
5ID9
 
 | | Crystal structure of equine serum albumin in complex with phosphorodithioate derivative of myristoyl cyclic phosphatidic acid (cPA) | | Descriptor: | (4S)-2-sulfanylidene-4-[(tetradecanoyloxy)methyl]-1,3,2lambda~5~-dioxaphospholane-2-thiolate, FORMIC ACID, MALONATE ION, ... | | Authors: | Sekula, B, Bujacz, A, Rytczak, P, Bujacz, G. | | Deposit date: | 2016-02-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural evidence of the species-dependent albumin binding of the modified cyclic phosphatidic acid with cytotoxic properties.
Biosci.Rep., 36, 2016
|
|
6NIC
 
 | | Crystal Structure of Medicago truncatula Agmatine Iminohydrolase (Deiminase) in Complex with 6-aminohexanamide | | Descriptor: | 1,2-ETHANEDIOL, 6-aminohexanamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Study of Agmatine Iminohydrolase FromMedicago truncatula, the Second Enzyme of the Agmatine Route of Putrescine Biosynthesis in Plants.
Front Plant Sci, 10, 2019
|
|
6O64
 
 | |
6WY2
 
 | | Crystal structure of RNA-10mer: CCGG(N4-methyl-C)GCCGG | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA-10mer: CCGG(4-methyl-C)GCCGG | | Authors: | Sekula, B, Ruszkowski, M, Mao, S, Haruehanroengra, P, Sheng, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
6OCI
 
 | | Crystal Structure of Equine Serum Albumin in Complex with Ibuprofen | | Descriptor: | FORMIC ACID, IBUPROFEN, SUCCINIC ACID, ... | | Authors: | Sekula, B, Zielinski, K, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-03-24 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural investigations of stereoselective profen binding by equine and leporine serum albumins.
Chirality, 32, 2020
|
|
6VCZ
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) | | Descriptor: | 2-METHOXYETHANOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6WY3
 
 | | Crystal structure of RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG; P212121 form | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*(LV2)P*GP*CP*CP*GP*G)-3') | | Authors: | Sekula, B, Ruszkowski, M, Mao, S, Haruehanroengra, P, Sheng, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
6VD2
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with S-adenosylmethionine | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6VD1
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with S-adenosylmethionine and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
