1JV2
 
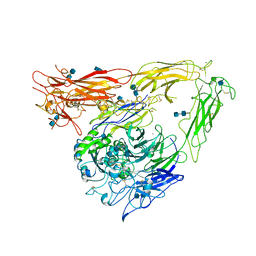 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHAVBETA3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.P, Stehle, T, Diefenbach, B, Zhang, R, Dunker, R, Scott, D, Joachimiak, A, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3.
Science, 294, 2001
|
|
7KWA
 
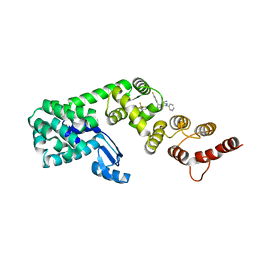 | | Structure of DCN1 bound to N-((4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-(trifluoromethyl)benzamide | | Descriptor: | Endolysin,DCN1-like protein 1, N-[(4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-(trifluoromethyl)benzamide | | Authors: | Kim, H.S, Hammill, J.T, Schulman, B.A, Guy, R.K, Scott, D.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Improvement of Oral Bioavailability of Pyrazolo-Pyridone Inhibitors of the Interaction of DCN1/2 and UBE2M.
J.Med.Chem., 64, 2021
|
|
5J4L
 
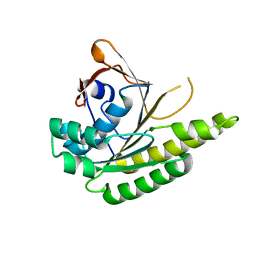 | | Apo-structure of humanised RadA-mutant humRadA22F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
1POB
 
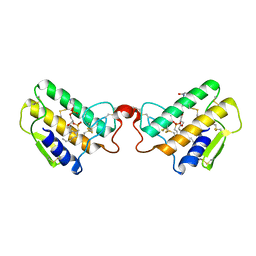 | | CRYSTAL STRUCTURE OF COBRA-VENOM PHOSPHOLIPASE A2 IN A COMPLEX WITH A TRANSITION-STATE ANALOGUE | | Descriptor: | 1-O-OCTYL-2-HEPTYLPHOSPHONYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | White, S.P, Scott, D.L, Otwinowski, Z, Sigler, P.B. | | Deposit date: | 1992-09-07 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of cobra-venom phospholipase A2 in a complex with a transition-state analogue.
Science, 250, 1990
|
|
8C1E
 
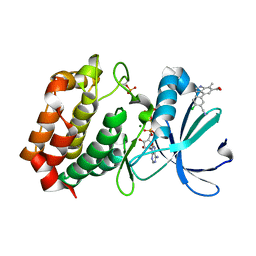 | | Aurora A kinase in complex with TPX2-inhibitor 9 | | Descriptor: | 4-(4-chloranyl-3-cyano-phenyl)-7-methyl-1~{H}-indole-6-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Fischer, G, Rocaboy, M, Blaszczyk, B, Moschetti, T, Wang, X, Scott, D.E, Coyne, A.G, Dagostin, C, Rooney, T, Bayly, A, Feng, J, Asteian, A, Alcaide-Lopez, A, Stockwell, S, Skidmore, J, Venkitaraman, A.R, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Selective inhibitors of the Aurora A-TPX2 protein-protein interaction exhibit in vivo efficacy as targeted anti-mitotic agent
To Be Published
|
|
3FN1
 
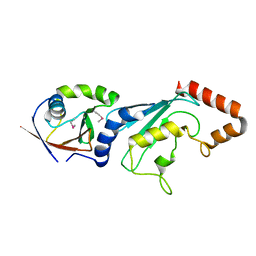 | | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. | | Descriptor: | NEDD8-activating enzyme E1 catalytic subunit, NEDD8-conjugating enzyme UBE2F | | Authors: | Huang, D.T, Ayrault, O, Hunt, H.W, Taherbhoy, A.M, Duda, D.M, Scott, D.C, Borg, L.A, Neale, G, Murray, P.J, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification
Mol.Cell, 33, 2009
|
|
5JEE
 
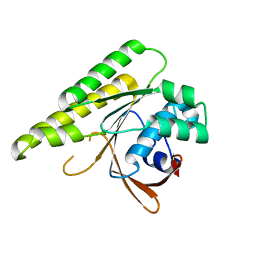 | | Apo-structure of humanised RadA-mutant humRadA26F | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
8C15
 
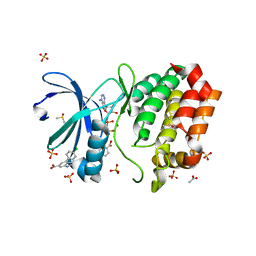 | | Aurora A kinase in complex with TPX2-inhibitor 3 | | Descriptor: | (1~{R},2~{R})-cyclohexane-1,2-dicarboxylic acid, 4-(4-chlorophenyl)-1~{H}-indole-6-carboxylic acid, ACETATE ION, ... | | Authors: | Fischer, G, Rocaboy, M, Blaszczyk, B, Moschetti, T, Wang, X, Scott, D.E, Coyne, A.G, Dagostin, C, Rooney, T, Bayly, A, Feng, J, Asteian, A, Alcaide-Lopez, A, Stockwell, S, Skidmore, J, Venkitaraman, A.R, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Selective inhibitors of the Aurora A-TPX2 protein-protein interaction exhibit in vivo efficacy as targeted anti-mitotic agents
To be published
|
|
6BG5
 
 | | Structure of 1-(benzo[d][1,3]dioxol-5-ylmethyl)-1-(1-propylpiperidin-4-yl)-3-(3-(trifluoromethyl)phenyl)urea bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-[(2H-1,3-benzodioxol-5-yl)methyl]-N-(1-propylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
6BG3
 
 | | Structure of (3S,4S)-1-benzyl-4-(3-(3-(trifluoromethyl)phenyl)ureido)piperidin-3-yl acetate bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-{(3S,4S)-1-benzyl-3-[(1S)-1-hydroxyethoxy]piperidin-4-yl}-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
1Y2O
 
 | | Structure of N-terminal domain IRSp53/BAIAP2 | | Descriptor: | BAI1-associated protein 2 isoform 1 | | Authors: | Millard, T.H, Bompard, G, Heung, M.-Y, Dafforn, T.R, Scott, D.J, Machesky, L.M, Futterer, K. | | Deposit date: | 2004-11-23 | | Release date: | 2005-02-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53
Embo J., 24, 2005
|
|
1DY3
 
 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | Deposit date: | 2000-01-21 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
3IOE
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.95 Ang resolution in complex with 5'-deoxy-5'-((R)-3,4-dihydroxybutylthio)-adenosine | | Descriptor: | 5'-S-[(3R)-3,4-dihydroxybutyl]-5'-thioadenosine, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Scott, D.E, Abell, C. | | Deposit date: | 2009-08-14 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Fragment-Based Approach to Probing Adenosine Recognition Sites by Using Dynamic Combinatorial Chemistry
Chembiochem, 10, 2009
|
|
3IOB
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.80 Ang resolution in complex with 5'-deoxy-5'-thioadenosine | | Descriptor: | 5'-thioadenosine, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Scott, D.E, Abell, C. | | Deposit date: | 2009-08-14 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Fragment-Based Approach to Probing Adenosine Recognition Sites by Using Dynamic Combinatorial Chemistry
Chembiochem, 10, 2009
|
|
3IOC
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 2.50 Ang resolution in complex with 5'-deoxy-5'-(benzyldisulfanyl)-adenosine | | Descriptor: | (2R,3R,4S,5S)-2-(6-amino-9H-purin-9-yl)-5-[(benzyldisulfanyl)methyl]tetrahydrofuran-3,4-diol, GLYCEROL, Pantothenate synthetase | | Authors: | Ciulli, A, Scott, D.E, Abell, C. | | Deposit date: | 2009-08-14 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Fragment-Based Approach to Probing Adenosine Recognition Sites by Using Dynamic Combinatorial Chemistry
Chembiochem, 10, 2009
|
|
3IOD
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.75 Ang resolution in complex with 5'-deoxy-5'-((3-nitrobenzyl)disulfanyl)-adenosine | | Descriptor: | (2R,3R,4S,5S)-2-(6-amino-9H-purin-9-yl)-5-{[(3-nitrobenzyl)disulfanyl]methyl}tetrahydrofuran-3,4-diol, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Scott, D.E, Abell, C. | | Deposit date: | 2009-08-14 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Fragment-Based Approach to Probing Adenosine Recognition Sites by Using Dynamic Combinatorial Chemistry
Chembiochem, 10, 2009
|
|
3RTR
 
 | | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Calabrese, M.F, Scott, D.C, Duda, D.M, Grace, C.R, Kurinov, I, Kriwacki, R.W, Schulman, B.A. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1DTO
 
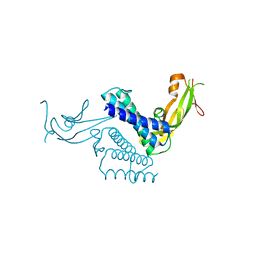 | | CRYSTAL STRUCTURE OF THE COMPLETE TRANSACTIVATION DOMAIN OF E2 PROTEIN FROM THE HUMAN PAPILLOMAVIRUS TYPE 16 | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Antson, A.A, Burns, J.E, Moroz, O.V, Scott, D.J, Sanders, C.M, Bronstein, I.B, Dodson, G.G, Wilson, K.S, Maitland, N. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Nature, 403, 2000
|
|
8BWV
 
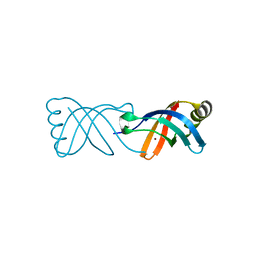 | |
1GUJ
 
 | | Insulin at pH 2: structural analysis of the conditions promoting insulin fibre formation. | | Descriptor: | INSULIN, SULFATE ION | | Authors: | Whittingham, J.L, Scott, D.J, Chance, K, Wilson, A, Finch, J, Brange, J, Dodson, G.G. | | Deposit date: | 2002-01-28 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Insulin at Ph2: Structural Analysis of the Conditions Promoting Insulin Fibre Formation
J.Mol.Biol., 318, 2002
|
|
1ODT
 
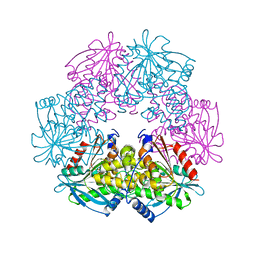 | | cephalosporin C deacetylase mutated, in complex with acetate | | Descriptor: | ACETATE ION, CEPHALOSPORIN C DEACETYLASE | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
2N5G
 
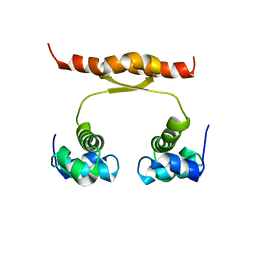 | | NMR structure of KorA, a plasmid-encoded, global transcription regulator KorA | | Descriptor: | TrfB transcriptional repressor protein | | Authors: | Rajasekar, K.V, Lovering, A.L, Dancea, F.V, Scott, D.J, Harris, S, Bingle, L.E, Roessle, M, Thomas, C.M, Hyde, E.I, White, S.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
1ODS
 
 | | Cephalosporin C deacetylase from Bacillus subtilis | | Descriptor: | CEPHALOSPORIN C DEACETYLASE, CHLORIDE ION, MAGNESIUM ION | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1LIJ
 
 | | STRUCTURE OF T. GONDII ADENOSINE KINASE BOUND TO PRODRUG 2 7-IODOTUBERCIDIN AND AMP-PCP | | Descriptor: | 2-RIBOFURANOSYL-3-IODO-2,3-DIHYDRO-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Scott, D.M, Mathews, I.I, Ealick, S.E, Roos, D.S, Ullman, B, Brennan, R.G. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of Toxoplasma gondii adenosine kinase reveal a novel catalytic mechanism and prodrug binding.
J.Mol.Biol., 298, 2000
|
|
1LIK
 
 | | STRUCTURE OF T. GONDII ADENOSINE KINASE BOUND TO ADENOSINE | | Descriptor: | ADENOSINE, CHLORIDE ION, SULFATE ION, ... | | Authors: | Schumacher, M.A, Scott, D.M, Mathews, I.I, Ealick, S.E, Roos, D.S, Ullman, B, Brennan, R.G. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of Toxoplasma gondii adenosine kinase reveal a novel catalytic mechanism and prodrug binding.
J.Mol.Biol., 298, 2000
|
|
