1DCS
 
 | | DEACETOXYCEPHALOSPORIN C SYNTHASE FROM S. CLAVULIGERUS | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE, SULFATE ION | | Authors: | Valegard, K, Terwisscha Van Scheltinga, A.C, Lloyd, M.D, Hara, T, Ramaswamy, S, Perrakis, A, Thompson, A, Lee, H.J, Baldwin, J.E, Schofield, C.J, Hajdu, J, Andersson, I. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a cephalosporin synthase.
Nature, 394, 1998
|
|
6ST3
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with 4-hydroxy-N-(4-phenoxybenzyl)-2-(1H-pyrazol-1-yl)pyrimidine-5-carboxamide | | Descriptor: | 4-oxidanyl-~{N}-[(4-phenoxyphenyl)methyl]-2-pyrazol-1-yl-pyrimidine-5-carboxamide, Egl nine homolog 1, FORMIC ACID, ... | | Authors: | Chowdhury, R, Holt-Martyn, J.P, Schofield, C.J. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.426 Å) | | Cite: | Structure-Activity Relationship and Crystallographic Studies on 4-Hydroxypyrimidine HIF Prolyl Hydroxylase Domain Inhibitors.
Chemmedchem, 15, 2020
|
|
2ILM
 
 | |
5A3U
 
 | | HIF prolyl hydroxylase 2 (PHD2/EGLN1) in complex with 6-(5-oxo-4-(1H- 1,2,3-triazol-1-yl)-2,5-dihydro-1H-pyrazol-1-yl)nicotinic acid | | Descriptor: | 6-(5-oxo-4-(1H-1,2,3-triazol-1-yl)-2,5-dihydro-1H-pyrazol-1-yl)nicotinic acid, EGL NINE HOMOLOG 1, MANGANESE (II) ION | | Authors: | Chowdhury, R, Gomez-Perez, V, Schofield, C.J. | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Potent and Selective Triazole-Based Inhibitors of the Hypoxia-Inducible Factor Prolyl-Hydroxylases with Activity in the Murine Brain.
Plos One, 10, 2015
|
|
8CF0
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with quinoxalinedione inhibitor H2 | | Descriptor: | 9-chloranyl-1,4-dihydropyrazino[2,3-c]quinoline-2,3-dione, DIMETHYL SULFOXIDE, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of human DNA cross-link repair 1A in complex with quinoxalinedione inhibitor H2
To Be Published
|
|
8CEW
 
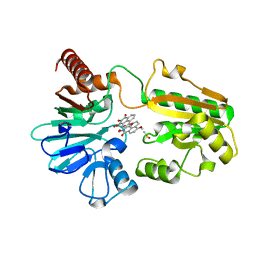 | | Crystal structure of human DNA cross-link repair 1A in complex with N-hydroxyimide inhibitor H1 | | Descriptor: | 6-methoxy-2-oxidanyl-benzo[de]isoquinoline-1,3-dione, DIMETHYL SULFOXIDE, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of human DNA cross-link repair 1A in complex with N-hydroxyimide inhibitor H1
To Be Published
|
|
8CG9
 
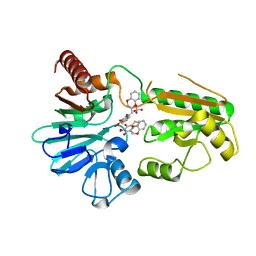 | | Crystal structure of human DNA cross-link repair 1A in complex with a cyclic N-hydroxyurea inhibitor | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, 1-[[(3~{R})-2,3-dihydro-1,4-benzodioxin-3-yl]methyl]-3-oxidanyl-thieno[3,2-d]pyrimidine-2,4-dione, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of human DNA cross-link repair 1A in complex with a cyclic N-hydroxyurea inhibitor
To Be Published
|
|
5N4S
 
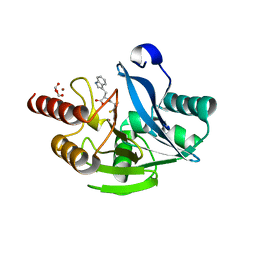 | | VIM-2 metallo-beta-lactamase in complex with ((S)-3-mercapto-2-methylpropanoyl)-D-tryptophan (Compound 3) | | Descriptor: | (2~{R})-3-(1~{H}-indol-3-yl)-2-[[(2~{S})-2-methyl-3-sulfanyl-propanoyl]amino]propanoic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
5N58
 
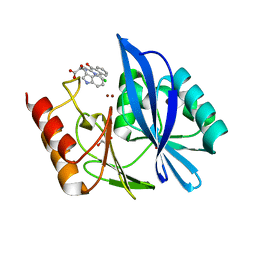 | | di-Zinc VIM-5 metallo-beta-lactamase in complex with (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-D-tryptophan (Compound 1) | | Descriptor: | (2~{R})-2-[(1-chloranyl-4-oxidanyl-isoquinolin-3-yl)carbonylamino]-3-(1~{H}-indol-3-yl)propanoic acid, Class B metallo-beta-lactamase, GLYCEROL, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
5N55
 
 | | mono-Zinc VIM-5 metallo-beta-lactamase in complex with (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-L-tryptophan (Compound 2) | | Descriptor: | (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-L-tryptophan, Class B metallo-beta-lactamase, ZINC ION | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
5NAI
 
 | | mono-Zinc VIM-5 metallo-beta-lactamase in complex with (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-D-tryptophan (Compound 1) | | Descriptor: | (2~{R})-2-[(1-chloranyl-4-oxidanyl-isoquinolin-3-yl)carbonylamino]-3-(1~{H}-indol-3-yl)propanoic acid, Class B metallo-beta-lactamase, FLUORIDE ION, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
5N4T
 
 | | VIM-2 metallo-beta-lactamase in complex with ((S)-3-mercapto-2-methylpropanoyl)-L-tryptophan (Compound 4) | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[[(2~{S})-2-methyl-3-sulfanyl-propanoyl]amino]propanoic acid, BENZOIC ACID, Beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
5NFO
 
 | |
5NFN
 
 | |
7P3L
 
 | |
7OR0
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
To Be Published
|
|
7OR1
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
TO BE PUBLISHED
|
|
5OP6
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc and GSK128863 | | Descriptor: | 2-[[1,3-dicyclohexyl-4-oxidanyl-2,6-bis(oxidanylidene)pyrimidin-5-yl]carbonylamino]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Leissing, T.M, Schofield, C.J, Clifton, I.J, Thinnes, C.C, Lu, X. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
5OPC
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc and Vadadustat | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Leissing, T.M, Schofield, C.J, Clifton, I.J, Lu, X, Zhang, D. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
7POY
 
 | | Spin labeled IPNS S55C variant in complex with Fe, ACV and NO | | Descriptor: | FE (III) ION, Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Clifton, I, Walla, C, Schofield, C.J. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Spectroscopic studies reveal details of substrate-induced conformational changes distant from the active site in isopenicillin N synthase.
J.Biol.Chem., 298, 2022
|
|
5T66
 
 | | Crystal Structure of CTX-M-15 with 1C | | Descriptor: | (3R)-3-(cyclohexylcarbonylamino)-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Beta-lactamase | | Authors: | Cahill, S.T, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Cyclic Boronates Inhibit All Classes of beta-Lactamases.
Antimicrob. Agents Chemother., 61, 2017
|
|
5OP8
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc and Molidustat | | Descriptor: | 2-(6-morpholin-4-ylpyrimidin-4-yl)-4-(1,2,3-triazol-1-yl)-1~{H}-pyrazol-3-one, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Leissing, T.M, Schofield, C.J, Clifton, I.J, Lu, X. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
7PJM
 
 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and Ca2+/2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
2BB4
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Asp-Phe at pH 5.0 | | Descriptor: | ASPARTIC ACID, CALCIUM ION, Elastase-1, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-17 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BD2
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Arg-Phe at pH 5.0 | | Descriptor: | ARGININE, CALCIUM ION, Elastase-1, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
