8BD6
 
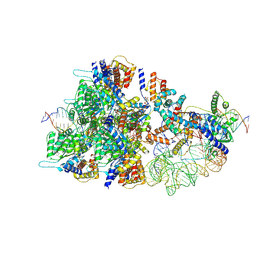 | | Cas12k-sgRNA-dsDNA-TnsC non-productive complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cas12k, DNA, ... | | Authors: | Schmitz, M, Querques, I, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
8BD5
 
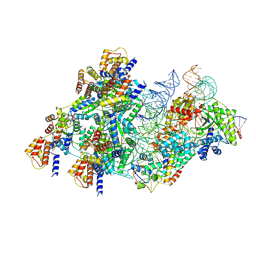 | | Cas12k-sgRNA-dsDNA-S15-TniQ-TnsC transposon recruitment complex | | Descriptor: | 30S ribosomal protein S15, ADENOSINE-5'-TRIPHOSPHATE, DNA non-target strand, ... | | Authors: | Schmitz, M, Querques, I, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
1XST
 
 | |
1XSG
 
 | |
1XSH
 
 | |
1XSU
 
 | |
7PLA
 
 | |
1F6X
 
 | |
1F6Z
 
 | |
1F78
 
 | |
1F79
 
 | |
1F7H
 
 | |
1F7F
 
 | |
1F7I
 
 | |
1F7G
 
 | |
2CD1
 
 | |
2CD6
 
 | |
2CD5
 
 | |
2CD3
 
 | |
8BD4
 
 | | TniQ-capped Tns-ATP-dsDNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*C)-3'), MAGNESIUM ION, ... | | Authors: | Querques, I, Schmitz, M, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
8P81
 
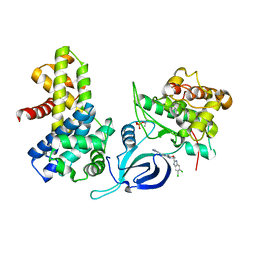 | | Crystal structure of human Cdk12/Cyclin K in complex with inhibitor SR-4835 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, ~{N}-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(1-methylpyrazol-4-yl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Anand, K, Schmitz, M, Geyer, M. | | Deposit date: | 2023-05-31 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The reversible inhibitor SR-4835 binds Cdk12/cyclin K in a noncanonical G-loop conformation.
J.Biol.Chem., 300, 2023
|
|
2LHT
 
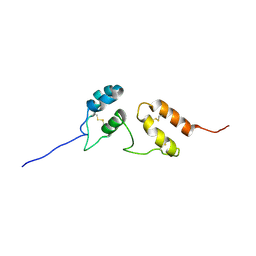 | | Solution structure of Venturia inaequalis cellophane-induced 1 protein (ViCin1) domains 1 and 2 | | Descriptor: | Cellophane-induced protein 1 | | Authors: | Mesarich, C.H, Schmitz, M, Tremouilhac, P, Greenwood, D.R, Mcgillivray, D.J, Templeton, M.D, Dingley, A.J. | | Deposit date: | 2011-08-16 | | Release date: | 2012-07-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and domain organization of the repeat protein Cin1 from the apple scab fungus.
Biochim.Biophys.Acta, 1824, 2012
|
|
