8THN
 
 | | KcsA M96V mutant with Y78ester in High K+ | | Descriptor: | KcsA Fab Heavy Chain, KcsA Fab Light Chain, POTASSIUM ION, ... | | Authors: | Reddi, R, Valiyaveetil, F.I. | | Deposit date: | 2023-07-17 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A facile approach for incorporating tyrosine esters to probe ion-binding sites and backbone hydrogen bonds.
J.Biol.Chem., 300, 2023
|
|
4OOK
 
 | | Third Metal bound M.tuberculosis methionine aminopeptidase | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase 2, SODIUM ION | | Authors: | Reddi, R, Addlagatta, A. | | Deposit date: | 2014-02-03 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the conserved active site cysteine of Mycobacterium tuberculosis methionine aminopeptidase with electrophilic reagents
Febs J., 281, 2014
|
|
4Q4E
 
 | | Crystal structure of E.coli aminopeptidase N in complex with actinonin | | Descriptor: | ACTINONIN, Aminopeptidase N, GLYCEROL, ... | | Authors: | Reddi, R, Ganji, R.J, Addlagatta, A. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of M1 family aminopeptidases by the natural product actinonin: Crystal structure in complex with E. coli aminopeptidase N.
Protein Sci., 24, 2015
|
|
4Q4I
 
 | | Crystal structure of E.coli aminopeptidase N in complex with amastatin | | Descriptor: | Amastatin, Aminopeptidase N, GLYCEROL, ... | | Authors: | Reddi, R, Ganji, R.J, Addlagatta, A. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the inhibition of M1 family aminopeptidases by the natural product actinonin: Crystal structure in complex with E. coli aminopeptidase N.
Protein Sci., 24, 2015
|
|
4X5K
 
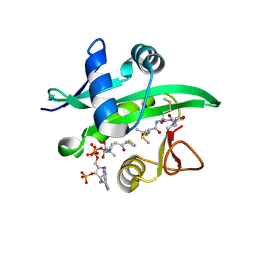 | |
7SIZ
 
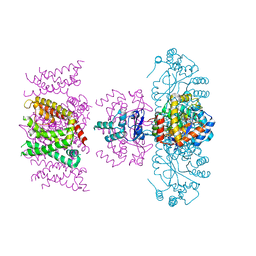 | | C-type inactivation in a voltage gated K+ channel | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Voltage gated potassium channel Kv1.2-Kv2.1, ... | | Authors: | Reddi, R, Riederer, E.A, Matulef, K, Whorton, M.R, Valiyaveetil, F.I. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for C-type inactivation in a Shaker family voltage-gated K + channel.
Sci Adv, 8, 2022
|
|
7RP0
 
 | | Structural Snapshots of Intermediates in the Gating of a K+ Channel | | Descriptor: | DIACYL GLYCEROL, KcsA Fab chain A, KcsA Fab chain B, ... | | Authors: | Reddi, R, Matulef, K, Riederer, E.A, Valiyaveetil, F.I. | | Deposit date: | 2021-08-02 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structures of Gating Intermediates in a K + channell.
J.Mol.Biol., 433, 2021
|
|
7SIT
 
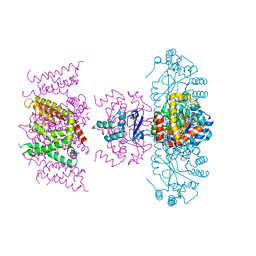 | | Crystal structure of Voltage gated potassium ion channel, Kv 1.2 chimera-3m | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXYGEN ATOM, POTASSIUM ION, ... | | Authors: | Reddi, R, Matulef, K, Riederer, E.A, Whorton, M.R, Valiyaveetil, F.I. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural basis for C-type inactivation in a Shaker family voltage-gated K + channel.
Sci Adv, 8, 2022
|
|
8DHR
 
 | | An ester mutant of SfGFP | | Descriptor: | Green fluorescent protein | | Authors: | Reddi, R, Valiyaveetil, F.I. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A facile approach for incorporating tyrosine esters to probe ion-binding sites and backbone hydrogen bonds.
J.Biol.Chem., 300, 2023
|
|
4IEC
 
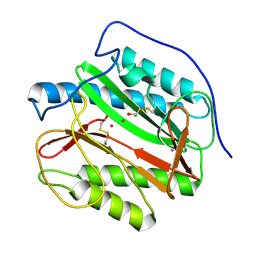 | | Cys105 covalent modification by 2-hydroxyethyl disulfide in Mycobacterium tuberculosis methionine aminopeptidase Type 1c | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase 2, POTASSIUM ION | | Authors: | Reddi, R, Gumpena, R, Kishor, C, Addlagatta, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-12-18 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective targeting of the conserved active site cysteine of Mycobacterium tuberculosis methionine aminopeptidase with electrophilic reagents
Febs J., 281, 2014
|
|
4IF7
 
 | | Mycobacterium Tuberculosis Methionine aminopeptidase Type 1c in complex with homocysteine-methyl disulfide | | Descriptor: | (2S)-2-amino-4-(methyldisulfanyl)butanoic acid, COBALT (II) ION, Methionine aminopeptidase 2 | | Authors: | Reddi, R, Gumpena, R, Kishor, C, Addlagatta, A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective targeting of the conserved active site cysteine of Mycobacterium tuberculosis methionine aminopeptidase with electrophilic reagents
Febs J., 281, 2014
|
|
4IDY
 
 | | Mycobacterium Tuberculosis Methionine aminopeptidase Type 1c in complex with 2-hydroxyethyl disulfide | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Methionine aminopeptidase 2, POTASSIUM ION | | Authors: | Reddi, R, Gumpena, R, Kishor, C, Addlagatta, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-12-18 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective targeting of the conserved active site cysteine of Mycobacterium tuberculosis methionine aminopeptidase with electrophilic reagents
Febs J., 281, 2014
|
|
3ROR
 
 | |
7M2H
 
 | |
7M2I
 
 | |
7M2J
 
 | |
3TB5
 
 | | Crystal Structure of the Enterococcus faecalis Methionine aminopeptidase apo form | | Descriptor: | CITRIC ACID, Methionine aminopeptidase | | Authors: | Kishor, C, Gumpena, R, Reddi, R, Addlagatta, A. | | Deposit date: | 2011-08-05 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of Enterococcus faecalis methionine aminopeptidase and design of microbe specific 2,2'-bipyridine based inhibitors
MEDCHEMCOMM, 3, 2012
|
|
5YQB
 
 | | Crystal structure of E.coli aminopeptidase N in complex with Puromycin | | Descriptor: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, Aminopeptidase N, GLYCEROL, ... | | Authors: | Marapaka, A.K, Ganji, R.J, Reddi, R, Addlagatta, A. | | Deposit date: | 2017-11-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Puromycin, a selective inhibitor of PSA acts as a substrate for other M1 family aminopeptidases: Biochemical and structural basis
Int.J.Biol.Macromol., 165, 2020
|
|
5YO1
 
 | | Structure of ePepN E298A mutant in complex with Puromycin | | Descriptor: | Aminopeptidase N, GLYCEROL, PUROMYCIN, ... | | Authors: | Ganji, R.J, Reddi, R, Marapaka, A.K, Addlagatta, A. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Puromycin, a selective inhibitor of PSA acts as a substrate for other M1 family aminopeptidases: Biochemical and structural basis
Int.J.Biol.Macromol., 165, 2020
|
|
5YQ1
 
 | | Crystal structure of E.coli aminopeptidase N in complex with O-Methyl-L-tyrosine | | Descriptor: | Aminopeptidase N, GLYCEROL, MALONATE ION, ... | | Authors: | Marapaka, A.K, Ganji, R.J, Reddi, R, Addlagatta, A. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of E.coli aminopeptidase N in complex with O-Methyl-L-tyrosine
To Be Published
|
|
5YQ2
 
 | | Crystal structure of E.coli aminopeptidase N in complex with Puromycin aminonucleoside | | Descriptor: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, Aminopeptidase N, GLYCEROL, ... | | Authors: | Marapaka, A.K, Ganji, R.J, Reddi, R, Addlagatta, A. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of E.coli aminopeptidase N in complex with Puromycin aminonucleoside
To Be Published
|
|
