5WME
 
 | | Crystal Structure of Amino Acids 1729-1786 of Human Beta Cardiac Myosin Fused to Gp7 as Anti-Parallel Four-Helix Bundle | | Descriptor: | Capsid assembly scaffolding protein,Myosin-7 | | Authors: | Andreas, M.P, Ajay, G, Gellings, J, Rayment, I. | | Deposit date: | 2017-07-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design considerations in coiled-coil fusion constructs for the structural determination of a problematic region of the human cardiac myosin rod.
J. Struct. Biol., 200, 2017
|
|
5WJB
 
 | | Crystal Structure of Amino Acids 1733-1797 of Human Beta Cardiac Myosin Fused to Gp7 | | Descriptor: | Capsid assembly scaffolding protein,Myosin-7 | | Authors: | Andreas, M.P, Ajay, G, Gellings, J, Rayment, I. | | Deposit date: | 2017-07-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Design considerations in coiled-coil fusion constructs for the structural determination of a problematic region of the human cardiac myosin rod.
J. Struct. Biol., 200, 2017
|
|
5TMD
 
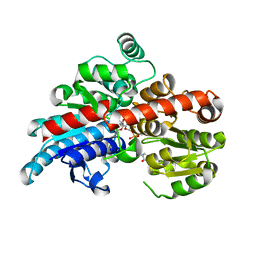 | | Crystal structure of Os79 from O. sativa in complex with U2F and trichothecene. | | Descriptor: | Glycosyltransferase, Os79, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE, ... | | Authors: | Wetterhorn, K, Newmister, S.A, Caniza, R.K, Busman, M, McCormick, S.P, Berthiller, F, Adam, G, Rayment, I. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol.
Biochemistry, 55, 2016
|
|
5WJ7
 
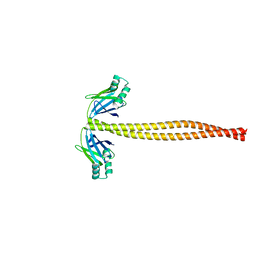 | | Crystal Structure of Amino Acids 1733-1797 of Human Beta Cardiac Myosin Fused to Xrcc4 | | Descriptor: | DNA repair protein XRCC4,Myosin-7 | | Authors: | Andreas, M.P, Ajay, G, Gellings, J, Rayment, I. | | Deposit date: | 2017-07-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design considerations in coiled-coil fusion constructs for the structural determination of a problematic region of the human cardiac myosin rod.
J. Struct. Biol., 200, 2017
|
|
1R6W
 
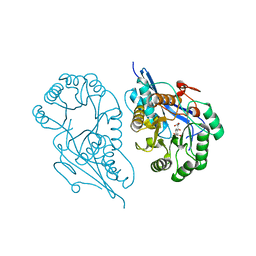 | | Crystal structure of the K133R mutant of o-Succinylbenzoate synthase (OSBS) from Escherichia coli. Complex with SHCHC | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, MAGNESIUM ION, o-Succinylbenzoate Synthase | | Authors: | Klenchin, V.A, Taylor Ringia, E.A, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-10-17 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural and Mutagenic Studies of the Mechanism of the Reaction Catalyzed by o-Succinylbenzoate Synthase from Escherichia coli
Biochemistry, 42, 2003
|
|
1SO6
 
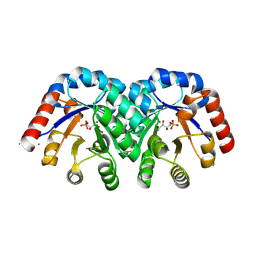 | | Crystal structure of E112Q/H136A double mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SO5
 
 | | Crystal structure of E112Q mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SO3
 
 | | Crystal structure of H136A mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
3GAH
 
 | | Structure of a F112H variant PduO-type ATP:corrinoid adenosyltransferase from Lactobacillus reuteri complexed with cobalamin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, Cobalamin adenosyltransferase PduO-like protein, ... | | Authors: | St Maurice, M, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2009-02-17 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Residue Phe112 of the human-type corrinoid adenosyltransferase (PduO) enzyme of Lactobacillus reuteri is critical to the formation of the four-coordinate Co(II) corrinoid substrate and to the activity of the enzyme.
Biochemistry, 48, 2009
|
|
3GAJ
 
 | | Structure of a C-terminal deletion variant of a PduO-type ATP:corrinoid adenosyltransferase from Lactobacillus reuteri complexed with cobalamin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, Cobalamin adenosyltransferase PduO-like protein, ... | | Authors: | St Maurice, M, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2009-02-17 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Residue Phe112 of the human-type corrinoid adenosyltransferase (PduO) enzyme of Lactobacillus reuteri is critical to the formation of the four-coordinate Co(II) corrinoid substrate and to the activity of the enzyme.
Biochemistry, 48, 2009
|
|
1SO4
 
 | | Crystal structure of K64A mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1FMV
 
 | | CRYSTAL STRUCTURE OF THE APO MOTOR DOMAIN OF DICTYOSTELLIUM MYOSIN II | | Descriptor: | CHLORIDE ION, MYOSIN II HEAVY CHAIN | | Authors: | Bauer, C.B, Holden, H.M, Thoden, J.B, Smith, R, Rayment, I. | | Deposit date: | 2000-08-18 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of the apo and MgATP-bound states of Dictyostelium discoideum myosin motor domain.
J.Biol.Chem., 275, 2000
|
|
1FMW
 
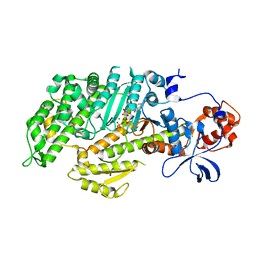 | | CRYSTAL STRUCTURE OF THE MGATP COMPLEX FOR THE MOTOR DOMAIN OF DICTYOSTELIUM MYOSIN II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MYOSIN II HEAVY CHAIN | | Authors: | Bauer, C.B, Holden, H.M, Thoden, J.B, Smith, R, Rayment, I. | | Deposit date: | 2000-08-18 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray structures of the apo and MgATP-bound states of Dictyostelium discoideum myosin motor domain.
J.Biol.Chem., 275, 2000
|
|
1FXA
 
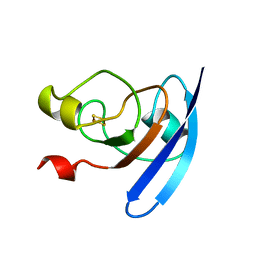 | | CRYSTALLIZATION AND STRUCTURE DETERMINATION TO 2.5-ANGSTROMS RESOLUTION OF THE OXIDIZED [2FE-2S] FERREDOXIN ISOLATED FROM ANABAENA 7120 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, [2FE-2S] FERREDOXIN | | Authors: | Rypniewski, W.R, Breiter, D.R, Benning, M.M, Wesenberg, G, Oh, B.-H, Markley, J.L, Rayment, I, Holden, H.M. | | Deposit date: | 1991-01-09 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and structure determination to 2.5-A resolution of the oxidized [2Fe-2S] ferredoxin isolated from Anabaena 7120.
Biochemistry, 30, 1991
|
|
1HPI
 
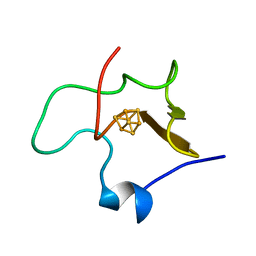 | | MOLECULAR STRUCTURE OF THE OXIDIZED HIGH-POTENTIAL IRON-SULFUR PROTEIN ISOLATED FROM ECTOTHIORHODOSPIRA VACUOLATA | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Benning, M.M, Meyer, T.E, Rayment, I, Holden, H.M. | | Deposit date: | 1993-12-09 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structure of the oxidized high-potential iron-sulfur protein isolated from Ectothiorhodospira vacuolata.
Biochemistry, 33, 1994
|
|
3FP0
 
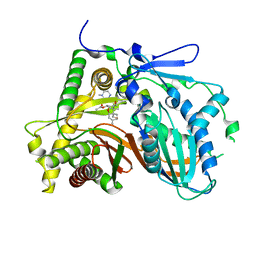 | | Structural and Functional Characterization of TRI3 Trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides | | Descriptor: | (3alpha)-15-hydroxy-12,13-epoxytrichothec-9-en-3-yl acetate, 15-O-acetyltransferase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Garvey, G.S, Rayment, I, McCormick, S.P, Alexander, N.J. | | Deposit date: | 2009-01-02 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of TRI3 trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides
Protein Sci., 18, 2009
|
|
1QZ5
 
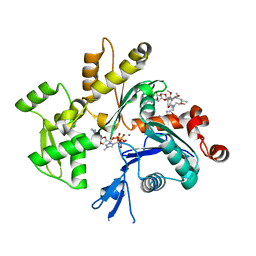 | | Structure of rabbit actin in complex with kabiramide C | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Klenchin, V.A, Allingham, J.S, King, R, Tanaka, J, Marriott, G, Rayment, I. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Trisoxazole macrolide toxins mimic the binding of actin-capping proteins to actin
Nat.Struct.Biol., 10, 2003
|
|
1QZ6
 
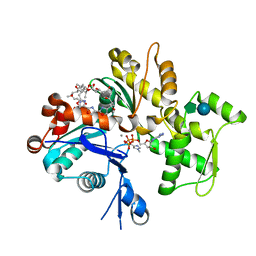 | | Structure of rabbit actin in complex with jaspisamide A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Klenchin, V.A, Allingham, J.S, King, R, Tanaka, J, Marriott, G, Rayment, I. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trisoxazole macrolide toxins mimic the binding of actin-capping proteins to actin
Nat.Struct.Biol., 10, 2003
|
|
1TQJ
 
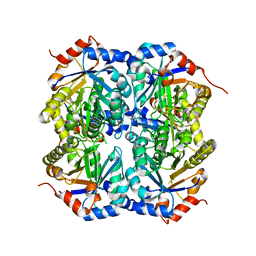 | | Crystal structure of D-ribulose 5-phosphate 3-epimerase from Synechocystis to 1.6 angstrom resolution | | Descriptor: | Ribulose-phosphate 3-epimerase | | Authors: | Wise, E.L, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-06-17 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of D-ribulose 5-phosphate 3-epimerase from Synechocystis to 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TKK
 
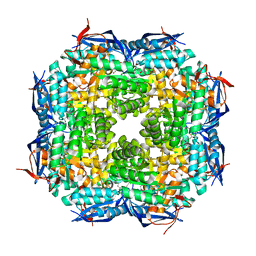 | | The Structure of a Substrate-Liganded Complex of the L-Ala-D/L-Glu Epimerase from Bacillus subtilis | | Descriptor: | ALANINE, GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Klenchin, V.A, Schmidt, D.M, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: Structure of a Substrate-Liganded Complex of the l-Ala-d/l-Glu Epimerase from Bacillus subtilis(,).
Biochemistry, 43, 2004
|
|
1PZ4
 
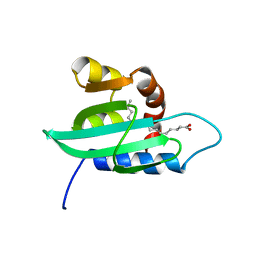 | | The structural determination of an insect (mosquito) Sterol Carrier Protein-2 with a ligand bound C16 Fatty Acid at 1.35 A resolution | | Descriptor: | PALMITIC ACID, sterol carrier protein 2 | | Authors: | Dyer, D.H, Lovell, S, Thoden, J.B, Holden, H.M, Rayment, I, Lan, Q. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Structural Determination of an Insect Sterol Carrier Protein-2 with a Ligand-bound C16 Fatty Acid at 1.35A Resolution
J.Biol.Chem., 278, 2003
|
|
1S22
 
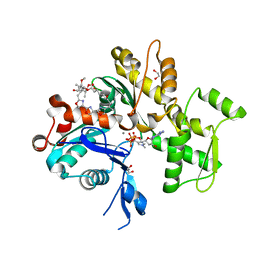 | | Absolute Stereochemistry of Ulapualide A | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Allingham, J.S, Tanaka, J, Marriott, G, Rayment, I. | | Deposit date: | 2004-01-07 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Absolute stereochemistry of ulapualide A
Org.Lett., 6, 2004
|
|
1PKN
 
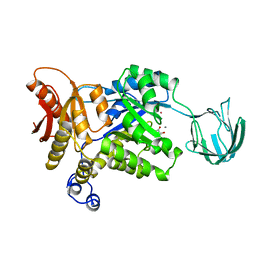 | | STRUCTURE OF RABBIT MUSCLE PYRUVATE KINASE COMPLEXED WITH MN2+, K+, AND PYRUVATE | | Descriptor: | MANGANESE (II) ION, POTASSIUM ION, PYRUVATE KINASE, ... | | Authors: | Larsen, T.M, Laughlin, L.T, Holden, H.M, Rayment, I, Reed, G.H. | | Deposit date: | 1994-03-25 | | Release date: | 1995-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of rabbit muscle pyruvate kinase complexed with Mn2+, K+, and pyruvate.
Biochemistry, 33, 1994
|
|
1Q6Q
 
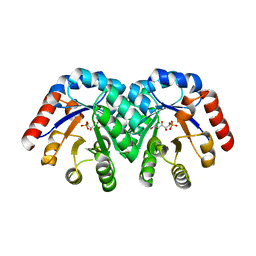 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound xylitol 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-XYLITOL 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
1Q6O
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-gulonaet 6-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-GULURONIC ACID 6-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
