3KXU
 
 | | Crystal structure of human ferritin FTL498InsTC pathogenic mutant | | Descriptor: | CADMIUM ION, Ferritin, SULFATE ION | | Authors: | Granier, T, Gallois, B, Langlois d'Estaintot, B, Arosio, P. | | Deposit date: | 2009-12-04 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mutant ferritin L-chains that cause neurodegeneration act in a dominant-negative manner to reduce ferritin iron incorporation.
J.Biol.Chem., 285, 2010
|
|
2F56
 
 | | Barnase cross-linked with glutaraldehyde soaked in 6M urea | | Descriptor: | Ribonuclease, UREA, ZINC ION | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-25 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
2F5M
 
 | | Cross-linked barnase soaked in bromo-ethanol | | Descriptor: | 2-BROMOETHANOL, Ribonuclease | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-26 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
2F4Y
 
 | | Barnase cross-linked with glutaraldehyde | | Descriptor: | Ribonuclease, ZINC ION | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-24 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
2F30
 
 | | Triclinic cross-linked Lysozyme soaked with 4.5M urea | | Descriptor: | Lysozyme C, NITRATE ION, UREA | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-18 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
2F4A
 
 | | Triclinic cross-linked lysozyme soaked with thiourea 1.5M | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION, ... | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-23 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
2F2N
 
 | | Triclinic hen egg lysozyme cross-linked by glutaraldehyde | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Prange, T, Salem, M, Mauguen, Y. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
2F4G
 
 | | Triclinic cross-linked lysozyme soaked in bromoethanol 1M | | Descriptor: | 2-BROMOETHANOL, Lysozyme C, NITRATE ION | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-23 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.654 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
6Q8S
 
 | |
6Q8R
 
 | |
6Q8Q
 
 | |
6QAR
 
 | |
8GQR
 
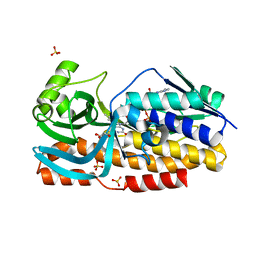 | | Crystal structure of VioD with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Ran, T, Wang, W, Xu, M. | | Deposit date: | 2022-08-30 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
4KNF
 
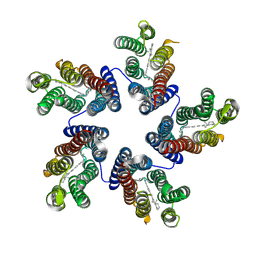 | | Crystal structure of a blue-light absorbing proteorhodopsin double-mutant D97N/Q105L from HOT75 | | Descriptor: | Blue-light absorbing proteorhodopsin, RETINAL | | Authors: | Ran, T, Ozorowski, G, Gao, Y, Wang, W, Luecke, H. | | Deposit date: | 2013-05-09 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KLY
 
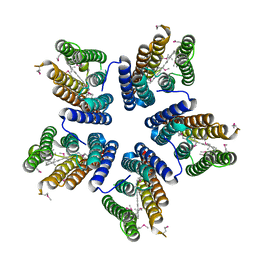 | | Crystal structure of a blue-light absorbing proteorhodopsin mutant D97N from HOT75 | | Descriptor: | Blue-light absorbing proteorhodopsin, RETINAL | | Authors: | Ran, T, Ozorowski, G, Gao, Y, Wang, W, Luecke, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1C1M
 
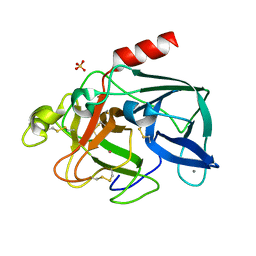 | | PORCINE ELASTASE UNDER XENON PRESSURE (8 BAR) | | Descriptor: | CALCIUM ION, PROTEIN (PORCINE ELASTASE), SULFATE ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
3L8W
 
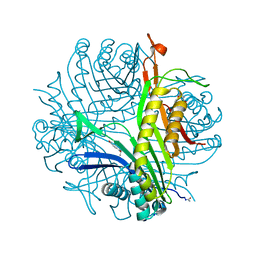 | | Urate oxidase from aspergillus flavus complexed with xanthin | | Descriptor: | SODIUM ION, Uricase, XANTHINE, ... | | Authors: | Prange, T, Gabison, L, Colloc'h, N, Chiadmi, M. | | Deposit date: | 2010-01-04 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Near-atomic resolution structures of urate oxidase complexed with its substrate and analogues: the protonation state of the ligand.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3L9G
 
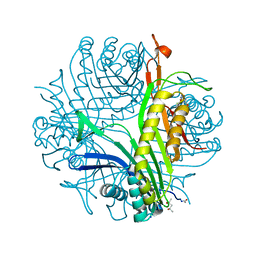 | | Urate oxidase complexed with uric acid and chloride | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Gabison, L. | | Deposit date: | 2010-01-05 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Near-atomic resolution structures of urate oxidase complexed with its substrate and analogues: the protonation state of the ligand.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1C3L
 
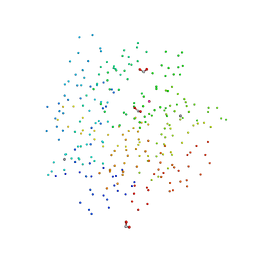 | | SUBTILISIN-CARLSBERG COMPLEXED WITH XENON (8 BAR) | | Descriptor: | CALCIUM ION, FORMIC ACID, SUBTILISIN-CARLSBERG, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
3LD4
 
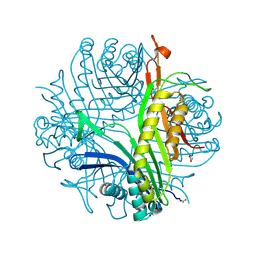 | | Urate oxidase complexed with 8-nitro xanthine | | Descriptor: | 1,2-ETHANEDIOL, 8-nitro-3,7-dihydro-1H-purine-2,6-dione, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Gabison, L. | | Deposit date: | 2010-01-12 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Near-atomic resolution structures of urate oxidase complexed with its substrate and analogues: the protonation state of the ligand.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3LBG
 
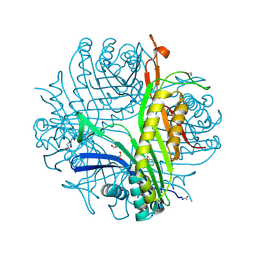 | | Urate oxidase complexed with 8-thio xanthine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-thioxo-3,7,8,9-tetrahydro-1H-purine-2,6-dione, ... | | Authors: | Prange, T, Colloc'h, N, Gabison, L. | | Deposit date: | 2010-01-08 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Near-atomic resolution structures of urate oxidase complexed with its substrate and analogues: the protonation state of the ligand.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6RGM
 
 | | urate oxidase under 130 bar of krypton | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-AZAXANTHINE, ACETATE ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2019-04-17 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
1C10
 
 | | CRYSTAL STRUCTURE OF HEW LYSOZYME UNDER PRESSURE OF XENON (8 BAR) | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-16 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
6R1Q
 
 | | murine Neuroglobin under 2 kBar of argon | | Descriptor: | ARGON, CHLORIDE ION, Neuroglobin, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P, Vallone, B. | | Deposit date: | 2019-03-14 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Argon-labelling protein crystals by high pressure cooling at 2000 Bar
To Be Published
|
|
