1Q8R
 
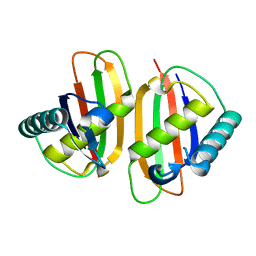 | | Structure of E.coli RusA Holliday junction resolvase | | Descriptor: | Crossover junction endodeoxyribonuclease rusA | | Authors: | Rafferty, J.B, Bolt, E.L, Muranova, T.A, Sedelnikova, S.E, Leonard, P, Pasquo, A, Baker, P.J, Rice, D.W, Sharples, G.J, Lloyd, R.G. | | Deposit date: | 2003-08-22 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The structure of Escherichia coli RusA endonuclease reveals a new Holliday junction DNA binding fold
Structure, 11, 2003
|
|
7QYP
 
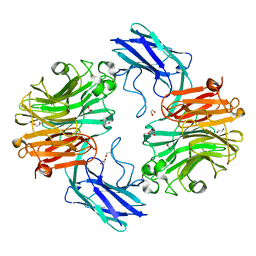 | | The structure of T. forsythia NanH | | Descriptor: | 1,2-ETHANEDIOL, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QY8
 
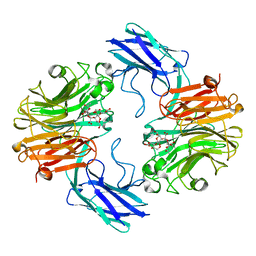 | | The structure of T. forsythia NanH | | Descriptor: | 3-sialyllactose, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QY9
 
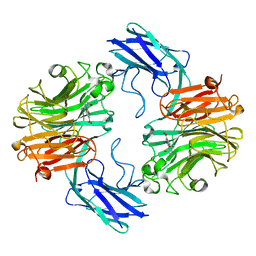 | | The structure of T.forsythia NanH with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QYJ
 
 | | The structure of T. forsythia NanH | | Descriptor: | BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QZ3
 
 | | The structure of T. forsythia NanH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QXO
 
 | | The structure of T. forsythia NanH | | Descriptor: | (2~{R},4~{S},5~{S},6~{S})-5-acetamido-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]-2-[[(2~{R},3~{R},4~{S},5~{R},6~{S})-3,4,5-tris(oxidanyl)-6-[(2~{R},3~{S},4~{R},5~{R})-1,2,4,5-tetrakis(oxidanyl)-6-oxidanylidene-hexan-3-yl]oxy-oxan-2-yl]methoxy]oxane-2-carboxylic acid, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
4L7T
 
 | | B. fragilis NanU | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, NanU sialic acid binding protein, ... | | Authors: | Rafferty, J.B, Phansopa, C, Stafford, G.P. | | Deposit date: | 2013-06-14 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species.
Biochem.J., 458, 2014
|
|
4KTW
 
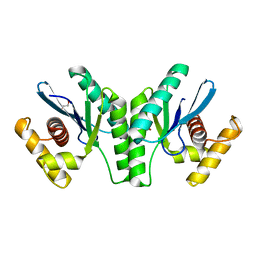 | | Lactococcus phage 67 RuvC - apo form | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, RuvC endonuclease | | Authors: | Rafferty, J.B, Green, V. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Mutants of phage bIL67 RuvC with enhanced Holliday junction binding selectivity and resolution symmetry.
Mol.Microbiol., 89, 2013
|
|
4KTZ
 
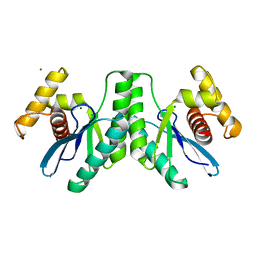 | | Lactococcus phage 67 RuvC | | Descriptor: | MAGNESIUM ION, RuvC endonuclease | | Authors: | Rafferty, J.B, Green, V. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutants of phage bIL67 RuvC with enhanced Holliday junction binding selectivity and resolution symmetry.
Mol.Microbiol., 89, 2013
|
|
1ENO
 
 | |
1ENP
 
 | |
1CUK
 
 | |
1CMB
 
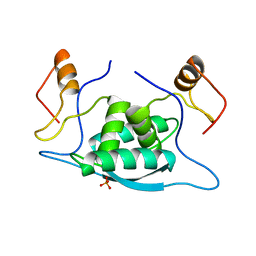 | |
5FDK
 
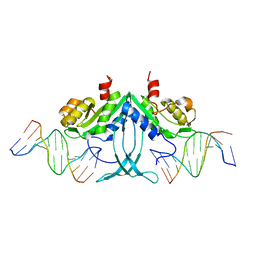 | |
7NSW
 
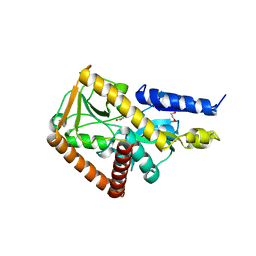 | | The structure of the SBP TarP_Csal in complex with coumarate | | Descriptor: | 1,2-ETHANEDIOL, 4'-HYDROXYCINNAMIC ACID, MAGNESIUM ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NR2
 
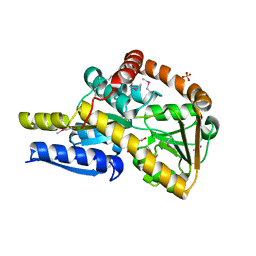 | | The structure of the SBP TarP_Sse in complex with coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NRR
 
 | | The structure of the SBP TarP_Csal in complex with caffeate | | Descriptor: | CAFFEIC ACID, MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-04 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NRA
 
 | | The structure of the SBP TarP_Sse in complex with cinnamate | | Descriptor: | HYDROCINNAMIC ACID, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NTD
 
 | | The structure of the SBP TarP_Csal in complex with ferulate | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NTE
 
 | | The structure of an open conformation of the SBP TarP_Csal | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
1MR9
 
 | | Crystal structure of Streptogramin A Acetyltransferase with acetyl-CoA bound | | Descriptor: | ACETYL COENZYME *A, Streptogramin A Acetyltransferase | | Authors: | Kehoe, L.E, Snidwongse, J, Courvalin, P, Rafferty, J.B, Murray, I.A. | | Deposit date: | 2002-09-18 | | Release date: | 2003-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Synercid (Quinupristin-Dalfopristin) Resistance in Gram-positive Bacterial Pathogens
J.Biol.Chem., 278, 2003
|
|
1MR7
 
 | | Crystal Structure of Streptogramin A Acetyltransferase | | Descriptor: | Streptogramin A Acetyltransferase | | Authors: | Kehoe, L.E, Snidwongse, J, Courvalin, P, Rafferty, J.B, Murray, I.A. | | Deposit date: | 2002-09-18 | | Release date: | 2003-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Synercid (Quinupristin-Dalfopristin) Resistance in Gram-positive Bacterial Pathogens
J.Biol.Chem., 278, 2003
|
|
1MRL
 
 | | Crystal structure of streptogramin A acetyltransferase with dalfopristin | | Descriptor: | 5-(2-DIETHYLAMINO-ETHANESULFONYL)-21-HYDROXY-10-ISOPROPYL-11,19-DIMETHYL-9,26-DIOXA-3,15,28-TRIAZA-TRICYCLO[23.2.1.00,255]OCTACOSA-1(27),12,17,19,25(28)-PENTAENE-2,8,14,23-TETRAONE, Streptogramin A acetyltransferase | | Authors: | Kehoe, L.E, Snidwongse, J, Courvalin, P, Rafferty, J.B, Murray, I.A. | | Deposit date: | 2002-09-18 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Synercid (Quinupristin-Dalfopristin) Resistance in Gram-positive Bacterial Pathogens
J.Biol.Chem., 278, 2003
|
|
7NQG
 
 | | The structure of the SBP TarP_Rhp in complex with 4-hydroxyphenylacetate | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENYLACETATE, PHOSPHATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
