6YZG
 
 | |
1YKI
 
 | | The structure of E. coli nitroreductase bound with the antibiotic nitrofurazone | | Descriptor: | CITRIC ACID, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
1YLU
 
 | | The structure of E. coli nitroreductase with bound acetate, crystal form 2 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
1YLR
 
 | | The structure of E.coli nitroreductase with bound acetate, crystal form 1 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
5MVI
 
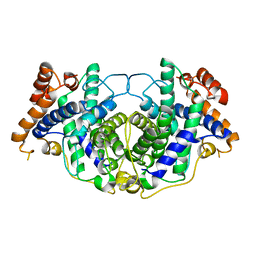 | |
5DYV
 
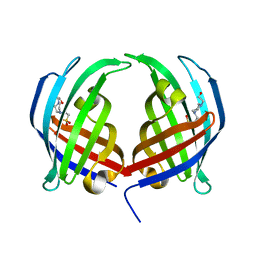 | | AbyU - wildtype | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, YD repeat-containing protein | | Authors: | Race, P.R, Byrne, M.J. | | Deposit date: | 2015-09-25 | | Release date: | 2016-06-08 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Catalytic Mechanism of a Natural Diels-Alderase Revealed in Molecular Detail.
J.Am.Chem.Soc., 138, 2016
|
|
5DZ7
 
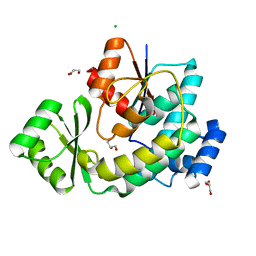 | |
4YZR
 
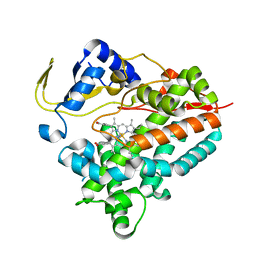 | |
4Z1P
 
 | | BspA_C_mut | | Descriptor: | Cell wall surface anchor protein | | Authors: | Race, P, Rego, S. | | Deposit date: | 2015-03-27 | | Release date: | 2016-06-29 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | BspA_C_mut
To Be Published
|
|
4Z23
 
 | | BspA_C_WT | | Descriptor: | 1,2-ETHANEDIOL, Cell wall surface anchor protein | | Authors: | Race, P, Rego, S. | | Deposit date: | 2015-03-28 | | Release date: | 2016-06-29 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | BspA_C_WT
To Be Published
|
|
1NWK
 
 | | CRYSTAL STRUCTURE OF MONOMERIC ACTIN IN THE ATP STATE | | Descriptor: | Actin, alpha skeletal muscle, CALCIUM ION, ... | | Authors: | Graceffa, P, Dominguez, R. | | Deposit date: | 2003-02-06 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics.
J.Biol.Chem., 278, 2003
|
|
7QU9
 
 | | Structure of aminodeoxychorismate synthase component 1 (PabB) from Bacillus subtilis spizizenii. | | Descriptor: | Anthranilate synthase component I family protein, GLYCEROL, TRYPTOPHAN | | Authors: | Rooms, L.D, Race, P.R, Back, C.B, Burton, N.B, Willis, C.L, Stach, J.E.M, Duke, P.W, Hawkins, C. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of aminodeoxychorismate synthase component 1 (PabB) from Bacillus subtilis spizizenii.
To Be Published
|
|
7QRH
 
 | |
7Z6P
 
 | | Diels-Alderase AbyU mutant - Y76F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, YD repeat-containing protein | | Authors: | Tiwari, K, Burton, N.M, Yang, S, Race, P.R. | | Deposit date: | 2022-03-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of Diels-Alderase AbyU mutant Y76F at 1.56 Angstroms resolution.
To Be Published
|
|
6YMN
 
 | |
5NO5
 
 | | AbyA5 Wildtype | | Descriptor: | AbyA5 | | Authors: | Byrne, M.J, Race, P.R. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Esterase-like Lyase Catalyzes Acetate Elimination in Spirotetronate/Spirotetramate Biosynthesis.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8OF7
 
 | | Cyc15 Diels Alderase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Back, C.R, Barringer, R.W.L, Zorn, K, Manzo-Ruiz, M, Race, P.R. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Interrogation of an Enzyme Library Reveals the Catalytic Plasticity of Naturally Evolved [4+2] Cyclases.
Chembiochem, 24, 2023
|
|
4YWF
 
 | | AbyA5 | | Descriptor: | AbyA5 | | Authors: | Byrne, M.J, Race, P.R. | | Deposit date: | 2015-03-20 | | Release date: | 2016-06-29 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Esterase-like Lyase Catalyzes Acetate Elimination in Spirotetronate/Spirotetramate Biosynthesis.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7QAN
 
 | | Cytochrome P450 Enzyme AbyV | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Parnell, A.E, Back, C.R, Race, P.R. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Role of Cytochrome P450 AbyV in the Final Stages of Abyssomicin C Biosynthesis.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4YX6
 
 | |
6FXD
 
 | | Crystal structure of MupZ from Pseudomonas fluorescens | | Descriptor: | MupZ | | Authors: | Wang, L, Parnell, A, Williams, C, Bakar, N.A, van der Kamp, M.W, Simpson, T.J, Race, P.R, Crump, M.P, Willis, C.L. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Rieske oxygenase/epoxide hydrolase-catalysed reaction cascade creates oxygen heterocycles in mupirocin biosynthesis
Nat Catal, 2018
|
|
7PI1
 
 | | Bacillus subtilis PabB | | Descriptor: | Aminodeoxychorismate synthase component 1, MAGNESIUM ION, TRYPTOPHAN | | Authors: | Rooms, L.D, Race, P.R. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-07 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Crystal structure of Bacillus subtilis PabB, component 1.
To be published
|
|
7PXO
 
 | | Structure of the Diels Alderase enzyme AbyU, from Micromonospora maris, co-crystallised with a non transformable substrate analogue | | Descriptor: | (2~{S},4~{S})-1-(4-methoxy-5-methyl-2-oxidanylidene-3~{H}-furan-3-yl)-2,4-dimethyl-dodecane-1,5-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, YD repeat-containing protein | | Authors: | Back, C.R, Race, P.R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Delineation of the Complete Reaction Cycle of a Natural Diels-Alderase
To Be Published
|
|
6SZC
 
 | |
2VY9
 
 | | Molecular architecture of the stressosome, a signal integration and transduction hub | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST | | Authors: | Marles-Wright, J, Grant, T, Delumeau, O, van Duinen, G, Firbank, S.J, Lewis, P.J, Murray, J.W, Newman, J.A, Quin, M.B, Race, P.R, Rohou, A, Tichelaar, W, van Heel, M, Lewis, R.J. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Architecture of the "Stressosome," a Signal Integration and Transduction Hub
Science, 322, 2008
|
|
