6OVC
 
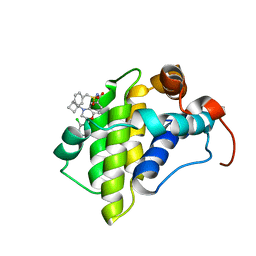 | | hMcl1 inhibitor complex | | Descriptor: | (2S)-N-(benzylsulfonyl)-4-(cyclobutylmethyl)-2-(2,4-dichlorophenyl)-3,4-dihydro-2H-1,4-benzoxazine-6-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Poppe, L. | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-22 | | Method: | SOLUTION NMR | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
7SUP
 
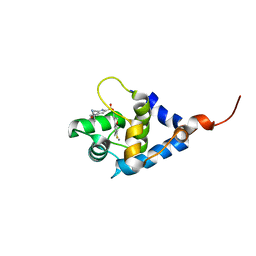 | | NMR structure of cTnC-TnI chimera bound to calcium and A1 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3S)-7-fluoro-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, CALCIUM ION, Troponin C, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SXC
 
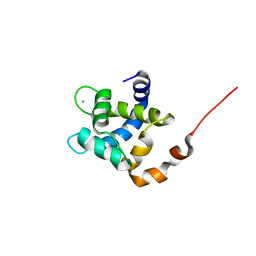 | | cTnC-TnI chimera complexed with calcium | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-22 | | Release date: | 2022-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SXD
 
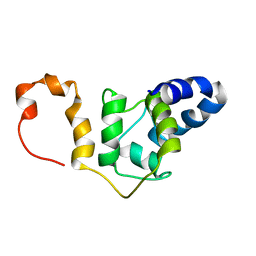 | | NMR solution structure TnC-TnI chimera | | Descriptor: | Troponin C, slow skeletal and cardiac muscles,Troponin I, cardiac muscle chimera | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-22 | | Release date: | 2022-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SWI
 
 | | cTnC-TnI chimera complexed with A2 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3R,4S)-7-fluoro-4-hydroxy-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SVC
 
 | | NMR structure of cTnC-TnI chimera bound to calcium and A2 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3R,4S)-7-fluoro-4-hydroxy-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, CALCIUM ION, Troponin C, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SWG
 
 | | cTnC-TnI chimera complexed with A1 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3S)-7-fluoro-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
2KEF
 
 | | Solution NMR structures of human hepcidin at 325K | | Descriptor: | Hepcidin | | Authors: | Jordan, J.B, Poppe, L, Hainu, M, Arvedson, T, Syed, R, Li, V, Kohno, H, Kim, H, Miranda, L.P, Cheetham, J, Sasu, B.J. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Hepcidin revisited, disulfide connectivity, dynamics, and structure.
J.Biol.Chem., 284, 2009
|
|
2MHS
 
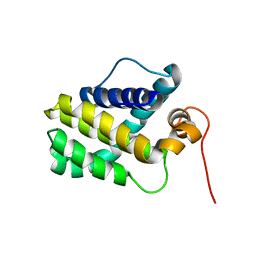 | | NMR Structure of human Mcl-1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Liu, G, Poppe, L, Aoki, K, Yamane, H, Lewis, J, Szyperski, T. | | Deposit date: | 2013-12-04 | | Release date: | 2014-05-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | High-Quality NMR Structure of Human Anti-Apoptotic Protein Domain Mcl-1(171-327) for Cancer Drug Design.
Plos One, 9, 2014
|
|
2LZG
 
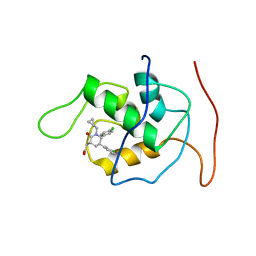 | | NMR Structure of Mdm2 (6-125) with Pip-1 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-(cyclopropylmethyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Michelsen, K.B, Jordan, J.B, Lewis, J, Long, A.M, Yang, E, Rew, Y, Zhou, J, Yakowec, P, Schnier, P.D, Huang, X, Poppe, L. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-07 | | Method: | SOLUTION NMR | | Cite: | Ordering of the N-Terminus of Human MDM2 by Small Molecule Inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
6HQF
 
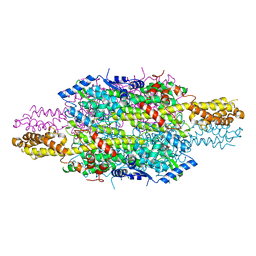 | | Structure of Phenylalanine ammonia-lyase from Petroselinum crispum in complex with (R)-APEP | | Descriptor: | Phenylalanine ammonia-lyase 1, [(1R)-1-amino-2-phenylethyl]phosphonic acid | | Authors: | Bata, Z, Molnar, B, Leveles, I, Poppe, L, Vertessy, G.B. | | Deposit date: | 2018-09-24 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Substrate Tunnel Engineering Aided by X-ray Crystallography and Functional Dynamics Swaps the Function of MIO-Enzymes
Acs Catalysis, 2021
|
|
6F6T
 
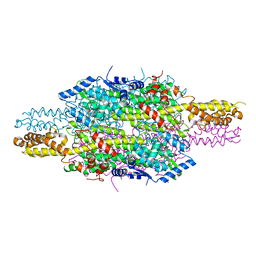 | |
6H2O
 
 | | APO structure of Phenylalanine ammonia-lyase from Petroselinum crispum | | Descriptor: | Phenylalanine ammonia-lyase 1 | | Authors: | Molnar, B, Bata, Z, Leveles, I, Poppe, L, Vertessy, G.B. | | Deposit date: | 2018-07-14 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Tunnel Engineering Aided by X-ray Crystallography and Functional Dynamics Swaps the Function of MIO-Enzymes
Acs Catalysis, 2021
|
|
