3S4L
 
 | | The CRISPR-associated Cas3 HD domain protein MJ0384 from Methanocaldococcus jannaschii | | Descriptor: | CALCIUM ION, CAS3 Metal dependent phosphohydrolase | | Authors: | Petit, P, Brown, G, Yakunin, A, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-19 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference.
Embo J., 30, 2011
|
|
3QYF
 
 | | Crystal structure of the CRISPR-associated protein SSO1393 from Sulfolobus solfataricus | | Descriptor: | CRISPR-ASSOCIATED PROTEIN, MAGNESIUM ION | | Authors: | Petit, P, Xu, X, Chang, C, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the CRISPR-associated protein SSO1393 from Sulfolobus solfataricus
To be Published
|
|
4XH9
 
 | | CRYSTAL STRUCTURE OF HUMAN RHOA IN COMPLEX WITH DH/PH FRAGMENT OF THE GUANINE NUCLEOTIDE EXCHANGE FACTOR NET1 | | Descriptor: | Neuroepithelial cell-transforming gene 1 protein, Transforming protein RhoA | | Authors: | Garcia, C, Petit, P, Boutin, J.A, Ferry, G, Vuillard, L. | | Deposit date: | 2015-01-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural study of the complex between neuroepithelial cell transforming gene 1 (Net1) and RhoA reveals a potential anticancer drug hot spot.
J. Biol. Chem., 293, 2018
|
|
3BXX
 
 | | Binding of two substrate analogue molecules to dihydroflavonol 4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dihydroflavonol 4-reductase | | Authors: | Trabelsi, N, Petit, P, Granier, T, Langlois d'Estaintot, B, Delrot, S, Gallois, B. | | Deposit date: | 2008-01-15 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols
Acta Crystallogr.,Sect.D, D64, 2008
|
|
3C1T
 
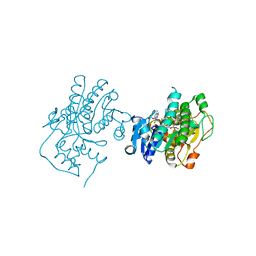 | | Binding of two substrate analogue molecules to dihydroflavonol 4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dihydroflavonol 4-reductase | | Authors: | Trabelsi, N, Petit, P, Granier, T, Langlois d'Estaintot, B, Delrot, S, Gallois, B. | | Deposit date: | 2008-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols
Acta Crystallogr.,Sect.D, D64, 2008
|
|
3R0P
 
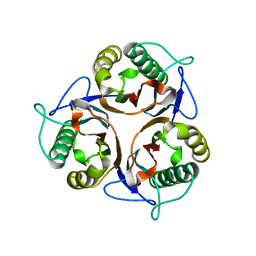 | | Crystal structure of L-PSP putative endoribonuclease from uncultured organism | | Descriptor: | L-PSP putative endoribonuclease | | Authors: | Cuff, M.E, Petit, P, Xu, X, Cui, H, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-08 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-PSP putative endoribonuclease from uncultured organism
To be Published
|
|
4K28
 
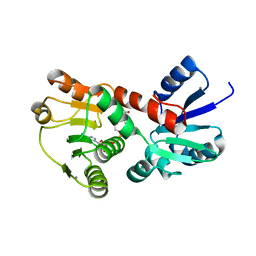 | | 2.15 Angstrom resolution crystal structure of a shikimate dehydrogenase family protein from Pseudomonas putida KT2440 in complex with NAD+ | | Descriptor: | MANGANESE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Garcia, C, Peek, J, Petit, P, Christendat, D. | | Deposit date: | 2013-04-08 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insights into the function of RifI2: structural and biochemical investigation of a new shikimate dehydrogenase family protein.
Biochim.Biophys.Acta, 1834, 2013
|
|
3V85
 
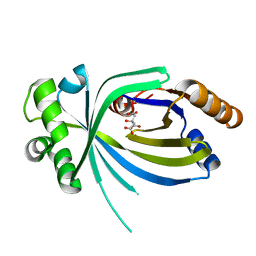 | |
3NWJ
 
 | |
