5TWD
 
 | | CTX-M-14 P167S apoenzyme | | Descriptor: | Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5TW6
 
 | | CTX-M-14 P167S:E166A mutant with acylated ceftazidime molecule | | Descriptor: | 1,2-ETHANEDIOL, ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-11 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5TWE
 
 | | CTX-M-14 P167S:S70G mutant enzyme crystallized with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
6CYQ
 
 | | Crystal structure of CTX-M-14 S70G/N106S beta-lactamase in complex with hydrolyzed cefotaxime | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro -2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, GLYCEROL | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
6CYU
 
 | | Crystal structure of CTX-M-14 S70G/N106S/D240G beta-lactamase in complex with hydrolyzed cefotaxime | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro -2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
6CYN
 
 | | CTX-M-14 N106S/D240G mutant | | Descriptor: | Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
6CYK
 
 | | CTX-M-14 N106S mutant | | Descriptor: | ACETATE ION, Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
5U53
 
 | | CTX-M-14 E166A with acylated ceftazidime molecule | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase, NITRATE ION | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-12-06 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
2KZA
 
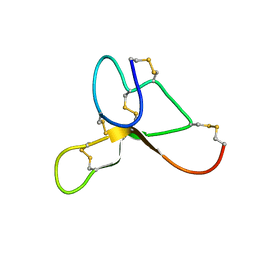 | | Solution structure of ASIP(80-132, P103A, P105A, Q115Y, S124Y) | | Descriptor: | Agouti-signaling protein | | Authors: | Patel, M.P, Cribb Fabersunne, C.S, Yang, Y, Kaelin, C.B, Barsh, G.S, Millhauser, G.L. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Loop-swapped chimeras of the agouti-related protein and the agouti signaling protein identify contacts required for melanocortin 1 receptor selectivity and antagonism.
J.Mol.Biol., 404, 2010
|
|
2L1J
 
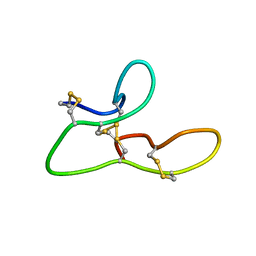 | | 1H assignments for ASIP(93-126, P103A, P105A, P111A, Q115Y, S124Y) | | Descriptor: | Agouti-signaling protein | | Authors: | Patel, M.P, Cribb Fabersunne, C.S, Yang, Y, Kaelin, C.B, Barsh, G.S, Millhauser, G.L. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Loop-swapped chimeras of the agouti-related protein and the agouti signaling protein identify contacts required for melanocortin 1 receptor selectivity and antagonism.
J.Mol.Biol., 404, 2010
|
|
3L4C
 
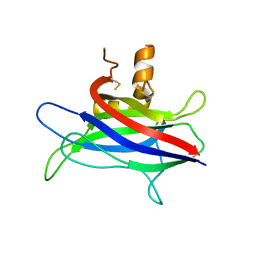 | | Structural basis of membrane-targeting by Dock180 | | Descriptor: | BETA-MERCAPTOETHANOL, Dedicator of cytokinesis protein 1 | | Authors: | Premkumar, L, Bobkov, A.A, Patel, M, Jaroszewski, L, Bankston, L.A, Stec, B, Vuori, K, Cote, J.-F, Liddington, R.C. | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of membrane targeting by the Dock180 family of Rho family guanine exchange factors (Rho-GEFs).
J.Biol.Chem., 285, 2010
|
|
3TUL
 
 | | Crystal structure of N-terminal region of Type III Secretion Major Translocator SipB (residues 82-226) | | Descriptor: | Cell invasion protein sipB | | Authors: | Barta, M.L, Dickenson, N.E, Patel, M, Keightley, J.A, Picking, W.D, Picking, W.L, Geisbrecht, B.V. | | Deposit date: | 2011-09-16 | | Release date: | 2012-02-15 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | The Structures of Coiled-Coil Domains from Type III Secretion System Translocators Reveal Homology to Pore-Forming Toxins.
J.Mol.Biol., 417, 2012
|
|
3IOX
 
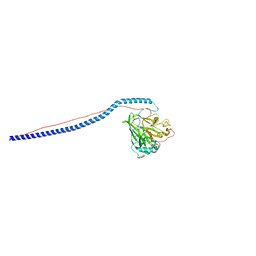 | | Crystal Structure of A3VP1 of AgI/II of Streptococcus mutans | | Descriptor: | AgI/II, CALCIUM ION, SULFITE ION, ... | | Authors: | Larson, M.R, Rajashankar, K.R, Patel, M, Robinette, R, Crowley, P, Michalek, S.M, Brady, L.J, Deivanayagam, C.C. | | Deposit date: | 2009-08-14 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IPK
 
 | | Crystal Structure of A3VP1 of AgI/II of Streptococcus mutans | | Descriptor: | AgI/II, CALCIUM ION, SULFATE ION, ... | | Authors: | Larson, M.R, Rajashankar, K.R, Patel, M, Robinette, R, Crowley, P, Michalek, S.M, Brady, L.J, Deivanayagam, C.C. | | Deposit date: | 2009-08-17 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7EWO
 
 | | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae | | Descriptor: | Cysteine synthase | | Authors: | Rahisuddin, R, Ekka, M.K, Singh, A.K, Saini, N, Patel, M, Kumar, N, Kumaran, S. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae
To Be Published
|
|
1ZY8
 
 | | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ciszak, E.M, Makal, A, Hong, Y.S, Vettaikkorumakankauv, A.K, Korotchkina, L.G, Patel, M.S. | | Deposit date: | 2005-06-09 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | How Dihydrolipoamide Dehydrogenase-binding Protein Binds Dihydrolipoamide Dehydrogenase in the Human Pyruvate Dehydrogenase Complex.
J.Biol.Chem., 281, 2006
|
|
1NI4
 
 | | HUMAN PYRUVATE DEHYDROGENASE | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component: Alpha subunit, ... | | Authors: | Ciszak, E, Korotchkina, L.G, Dominiak, P.M, Sidhu, S, Patel, M.S. | | Deposit date: | 2002-12-20 | | Release date: | 2003-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Flip-Flop Action of Thiamin Pyrophosphate-Dependent Enzymes Revealed by Human Pyruvate Dehydrogenase
J.Biol.Chem., 278, 2003
|
|
1TXT
 
 | | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, ACETOACETYL-COENZYME A | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
1TVZ
 
 | | Crystal structure of 3-hydroxy-3-methylglutaryl-coenzyme A synthase from Staphylococcus aureus | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, SULFATE ION | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
5VTH
 
 | | CTX-M-14 P167S:E166A mutant | | Descriptor: | Beta-lactamase | | Authors: | Hu, L, Patel, M, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2017-05-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
2KDI
 
 | | Solution structure of a Ubiquitin/UIM fusion protein | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein 27 fusion protein | | Authors: | Sgourakis, N.G, Patel, M.M, Garcia, A.E, Makhatadze, G.I, McCallum, S.A. | | Deposit date: | 2009-01-09 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Conformational Dynamics and Structural Plasticity Play Critical Roles in the Ubiquitin Recognition of a UIM Domain.
J.Mol.Biol., 396, 2010
|
|
3OAZ
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | CHLORIDE ION, Fab 2G12, heavy chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-06 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OB0
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | 7-deoxy-L-glycero-alpha-D-manno-heptopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, Fab 2G12, heavy chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-06 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OAY
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2VSZ
 
 | | Crystal Structure of the ELMO1 PH domain | | Descriptor: | ENGULFMENT AND CELL MOTILITY PROTEIN 1 | | Authors: | Komander, D, Patel, M, Barford, D, Cote, J.-F. | | Deposit date: | 2008-05-01 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Alpha-Helical Extension of the Elmo1 Pleckstrin Homology Domain Mediates Direct Interaction to Dock180 and is Critical in Rac Signaling.
Molecular Biology of the Cell, 19, 2008
|
|
