1TNN
 
 | |
1LY7
 
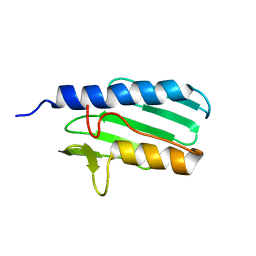 | | The solution structure of the the c-terminal domain of frataxin, the protein responsible for friedreich ataxia | | Descriptor: | frataxin | | Authors: | Musco, G, Stier, G, Kolmerer, B, Adinolfi, S, Martin, S, Frenkiel, T, Gibson, T, Pastore, A. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-26 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Towards a structural understanding of Friedreich's
ataxia: the solution structure of frataxin
Structure Fold.Des., 8, 2000
|
|
1TIT
 
 | | TITIN, IG REPEAT 27, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TITIN, I27 | | Authors: | Improta, S, Politou, A.S, Pastore, A. | | Deposit date: | 1996-02-02 | | Release date: | 1996-07-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity.
Structure, 4, 1996
|
|
1TIU
 
 | | TITIN, IG REPEAT 27, NMR, 24 STRUCTURES | | Descriptor: | TITIN, I27 | | Authors: | Improta, S, Politou, A.S, Pastore, A. | | Deposit date: | 1996-02-02 | | Release date: | 1996-07-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity.
Structure, 4, 1996
|
|
5ONK
 
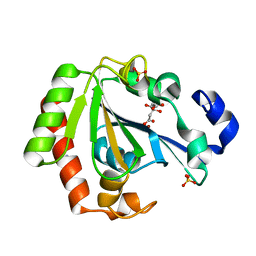 | | Native YndL | | Descriptor: | CITRATE ANION, IMIDAZOLE, SULFATE ION, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
5ONL
 
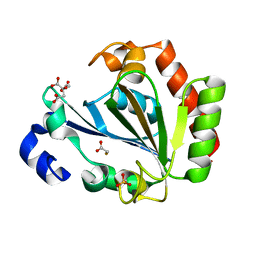 | | YNDL-apo (Zinc-free) | | Descriptor: | CITRATE ANION, SULFATE ION, YndL, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
Febs J., 285, 2018
|
|
1RGW
 
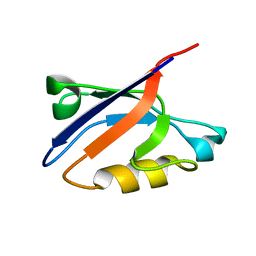 | | Solution Structure of ZASP's PDZ domain | | Descriptor: | ZASP protein | | Authors: | Au, Y, Atkinson, R.A, Pallavicini, A, Joseph, C, Martin, S.R, Muskett, F.W, Guerrini, R, Faulkner, G, Pastore, A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of ZASP PDZ Domain; Implications for Sarcomere Ultrastructure and Enigma Family Redundancy.
Structure, 12, 2004
|
|
2BKD
 
 | | Structure of the N-terminal domain of Fragile X Mental Retardation Protein | | Descriptor: | Fragile X messenger ribonucleoprotein 1 | | Authors: | Ramos, A, Hollingworth, D, Adinolfi, S, Castets, M, Kelly, G, Frenkiel, T.A, Bardoni, B, Pastore, A. | | Deposit date: | 2005-02-15 | | Release date: | 2006-01-18 | | Last modified: | 2023-02-01 | | Method: | SOLUTION NMR | | Cite: | The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction.
Structure, 14, 2006
|
|
2FMR
 
 | | KH1 FROM THE FRAGILE X PROTEIN FMR1, NMR, 18 STRUCTURES | | Descriptor: | FMR1 PROTEIN | | Authors: | Musco, G, Kharrat, A, Stier, G, Fraternali, F, Gibson, T.J, Nilges, M, Pastore, A. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome.
Nat.Struct.Biol., 4, 1997
|
|
1NEB
 
 | |
2JQD
 
 | |
1RPR
 
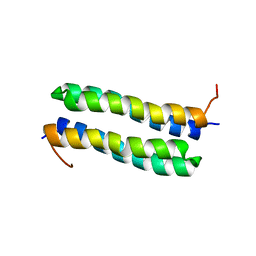 | | THE STRUCTURE OF COLE1 ROP IN SOLUTION | | Descriptor: | ROP | | Authors: | Eberle, W, Pastore, A, Klaus, W, Sander, C, Roesch, P. | | Deposit date: | 1991-10-09 | | Release date: | 1994-01-31 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of ColE1 rop in solution.
J.Biomol.NMR, 1, 1991
|
|
6FCO
 
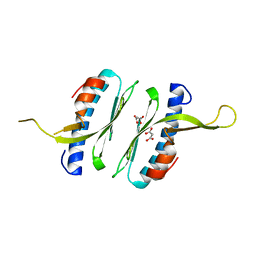 | | Structural and functional characterisation of Frataxin (FXN) like protein from Chaetomium thermophilum | | Descriptor: | MALONIC ACID, Mitochondrial frataxin-like protein | | Authors: | Jamshidiha, M, Rasheed, M, Pastore, A, Cota, E. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and functional characterization of a frataxin from a thermophilic organism.
FEBS J., 286, 2019
|
|
2BTT
 
 | |
2JRI
 
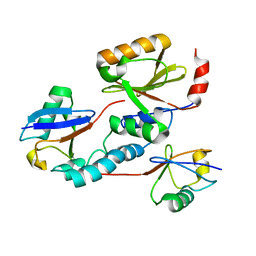 | | Solution structure of the Josephin domain of Ataxin-3 in complex with ubiquitin molecule. | | Descriptor: | Ataxin-3, UBC protein | | Authors: | Nicastro, G, Masino, L, Esposito, V, Menon, R, Pastore, A. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Understanding the plasticity of the ubiquitin-protein recognition code: the josephin domain of ataxin-3 is a diubiquitin binding motif
To be Published
|
|
2M41
 
 | |
2JNF
 
 | |
2LFU
 
 | | The structure of a N. meningitides protein targeted for vaccine development | | Descriptor: | Gna2132 | | Authors: | Esposito, V, Musi, V, De Chiara, C, Kelly, G, Veggi, D, Pizza, M, Pastore, A. | | Deposit date: | 2011-07-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal Domain of Neisseria Heparin Binding Antigen (NHBA), One of the Main Antigens of a Novel Vaccine against Neisseria meningitidis.
J.Biol.Chem., 286, 2011
|
|
